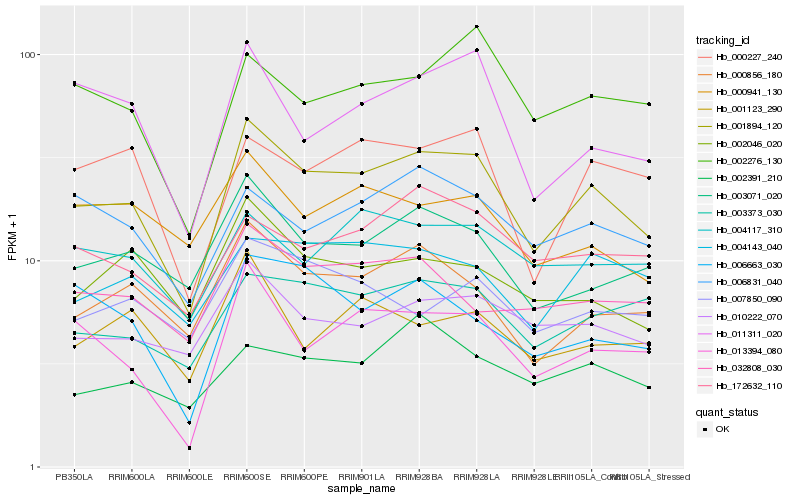
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001894_120 |
0.0 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 2 |
Hb_000856_180 |
0.109764634 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 3 |
Hb_003071_020 |
0.1148332838 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 4 |
Hb_013394_080 |
0.1162375042 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1D isoform X2 [Jatropha curcas] |
| 5 |
Hb_010222_070 |
0.1181439543 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_003373_030 |
0.1204203837 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATG1 [Jatropha curcas] |
| 7 |
Hb_011311_020 |
0.124088839 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000227_240 |
0.1250503593 |
- |
- |
hypothetical protein JCGZ_02139 [Jatropha curcas] |
| 9 |
Hb_006831_040 |
0.1257521887 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004143_040 |
0.1258902622 |
- |
- |
argininosuccinate lyase, putative [Ricinus communis] |
| 11 |
Hb_002276_130 |
0.1261491462 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 12 |
Hb_002046_020 |
0.1270462322 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 13 |
Hb_032808_030 |
0.1279790858 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_004117_310 |
0.1291301769 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 15 |
Hb_172632_110 |
0.1297475368 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 16 |
Hb_006663_030 |
0.1309872938 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_002391_210 |
0.1324926615 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 18 |
Hb_007850_090 |
0.1349351792 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001123_290 |
0.1349594356 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 20 |
Hb_000941_130 |
0.1351488812 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |