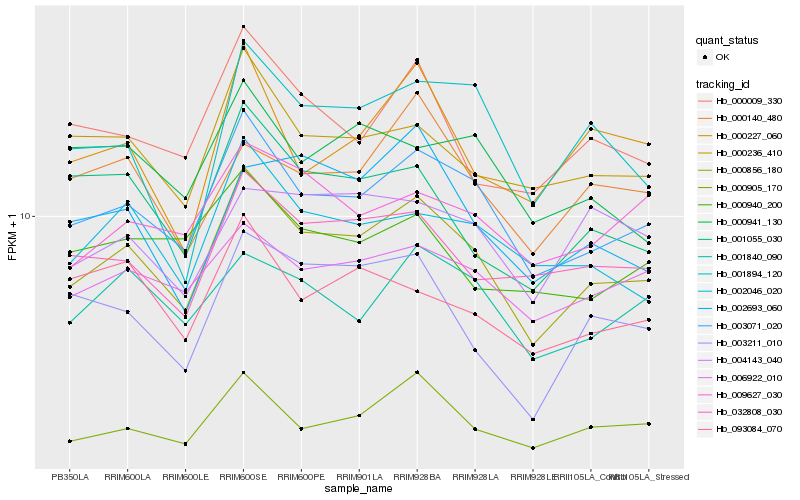
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_180 |
0.0 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 2 |
Hb_003071_020 |
0.052114681 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 3 |
Hb_032808_030 |
0.100713715 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_002046_020 |
0.101890364 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 5 |
Hb_000905_170 |
0.1019408865 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 6 |
Hb_000236_410 |
0.104698625 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 7 |
Hb_000009_330 |
0.1050596285 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_006922_010 |
0.105180041 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 9 |
Hb_001840_090 |
0.1093738836 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_001894_120 |
0.109764634 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 11 |
Hb_001055_030 |
0.1100332435 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003211_010 |
0.1148451488 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 13 |
Hb_009627_030 |
0.1153955926 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 14 |
Hb_000227_060 |
0.1177158612 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 15 |
Hb_000140_480 |
0.1199768117 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 16 |
Hb_002693_060 |
0.1215493305 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000941_130 |
0.1226351727 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 18 |
Hb_093084_070 |
0.1229328928 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 19 |
Hb_004143_040 |
0.1230251502 |
- |
- |
argininosuccinate lyase, putative [Ricinus communis] |
| 20 |
Hb_000940_200 |
0.1230467873 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |