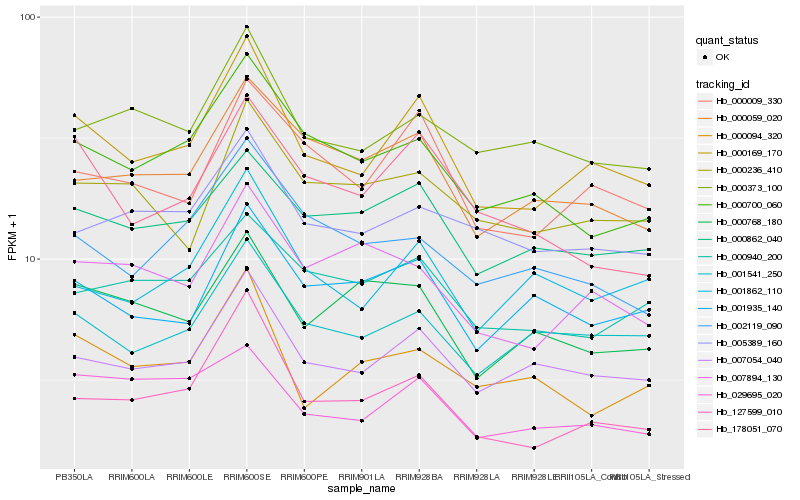
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000169_170 |
0.0 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 2 |
Hb_007054_040 |
0.0754944875 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 3 |
Hb_000009_330 |
0.0820062374 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_001541_250 |
0.0906375659 |
- |
- |
PREDICTED: integrator complex subunit 9 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_127599_010 |
0.0936943277 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 6 |
Hb_000862_040 |
0.0964729819 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 7 |
Hb_001862_110 |
0.1016926278 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 8 |
Hb_000700_060 |
0.1018389442 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 9 |
Hb_000373_100 |
0.1043817158 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 10 |
Hb_000059_020 |
0.1046467623 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 11 |
Hb_001935_140 |
0.1048468509 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 12 |
Hb_029695_020 |
0.1052341902 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_178051_070 |
0.1072055432 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 14 |
Hb_005389_160 |
0.1093151304 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 15 |
Hb_000940_200 |
0.1102126277 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 16 |
Hb_000236_410 |
0.1116220872 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 17 |
Hb_007894_130 |
0.1138979327 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 18 |
Hb_000094_320 |
0.1147863965 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 19 |
Hb_002119_090 |
0.1149896587 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 20 |
Hb_000768_180 |
0.1157784672 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |