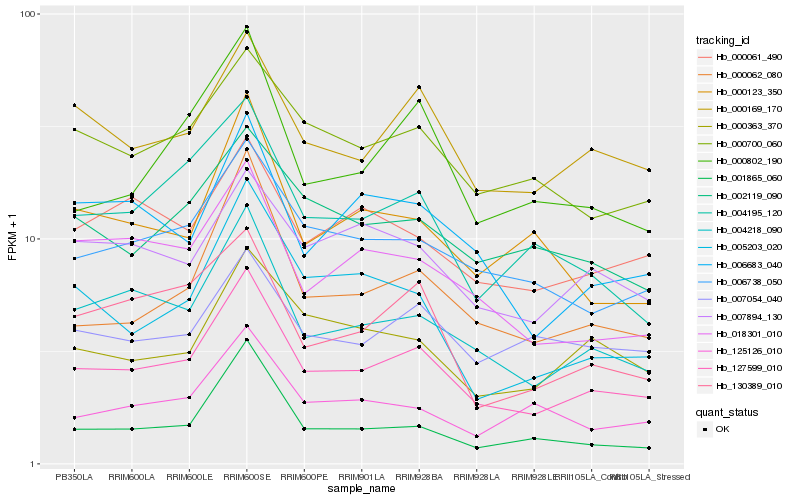
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127599_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 2 |
Hb_004218_090 |
0.0772548417 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000062_080 |
0.0935141085 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 4 |
Hb_000169_170 |
0.0936943277 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 5 |
Hb_006738_050 |
0.0969110683 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 6 |
Hb_000700_060 |
0.1070148592 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 7 |
Hb_006683_040 |
0.1088712058 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 8 |
Hb_018301_010 |
0.1095032944 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 9 |
Hb_000802_190 |
0.1097485387 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 10 |
Hb_000363_370 |
0.1102273014 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 11 |
Hb_004195_120 |
0.1125218215 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 12 |
Hb_130389_010 |
0.1129422741 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 13 |
Hb_007894_130 |
0.1156942166 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 14 |
Hb_000061_490 |
0.1157276758 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 15 |
Hb_005203_020 |
0.1163661995 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 16 |
Hb_002119_090 |
0.1166328752 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 17 |
Hb_007054_040 |
0.1173912474 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 18 |
Hb_000123_350 |
0.1178036854 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_001865_060 |
0.1190861401 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 20 |
Hb_125126_010 |
0.1198491895 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |