| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
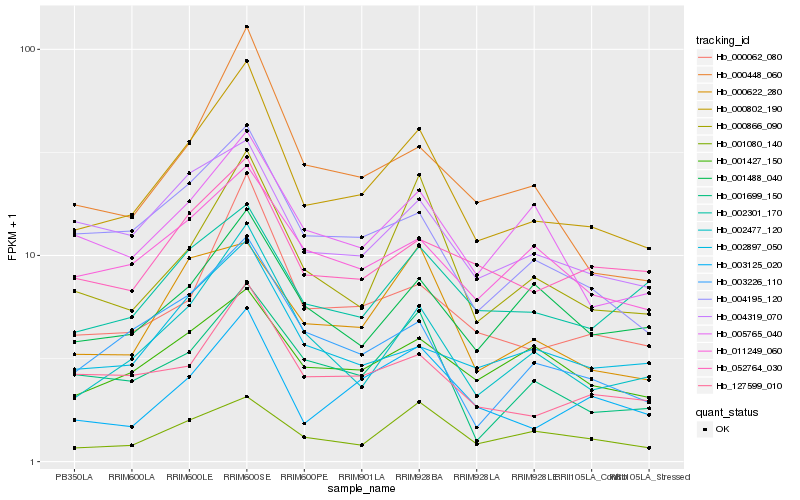
Hb_000802_190 |
0.0 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 2 |
Hb_000062_080 |
0.1053304058 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 3 |
Hb_002477_120 |
0.1054981339 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_127599_010 |
0.1097485387 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 5 |
Hb_000866_090 |
0.1100570177 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_004195_120 |
0.1134696586 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_003125_020 |
0.1189669416 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 8 |
Hb_000448_060 |
0.1209757369 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 9 |
Hb_004319_070 |
0.1224062444 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 10 |
Hb_002897_050 |
0.1226188112 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 11 |
Hb_003226_110 |
0.1233454437 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 12 |
Hb_052764_030 |
0.1240290572 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 13 |
Hb_001427_150 |
0.12542374 |
- |
- |
PREDICTED: uncharacterized protein LOC105637790 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000622_280 |
0.1256487384 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011249_060 |
0.1271450809 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 16 |
Hb_001699_150 |
0.1275056008 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 17 |
Hb_002301_170 |
0.128363448 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |
| 18 |
Hb_001080_140 |
0.1285128837 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005765_040 |
0.129483814 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 20 |
Hb_001488_040 |
0.129762859 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |