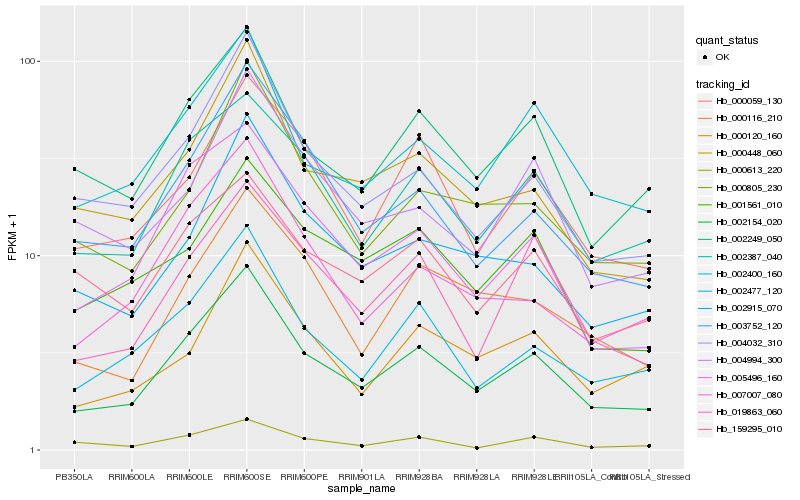
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002154_020 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_002387_040 |
0.0932359223 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 3 |
Hb_002249_050 |
0.094841164 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 4 |
Hb_002477_120 |
0.0950629479 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 5 |
Hb_002400_160 |
0.0958120468 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 6 |
Hb_004032_310 |
0.1032168424 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_019863_060 |
0.1032447511 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 8 |
Hb_005496_160 |
0.1033924012 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000448_060 |
0.1044966789 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 10 |
Hb_003752_120 |
0.1075407103 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000059_130 |
0.107822413 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 12 |
Hb_007007_080 |
0.113455003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000116_210 |
0.1142476071 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 14 |
Hb_000805_230 |
0.1142674812 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 15 |
Hb_002915_070 |
0.1156821599 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 16 |
Hb_001561_010 |
0.1194892278 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000120_160 |
0.1195931748 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000613_220 |
0.1220637153 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 19 |
Hb_004994_300 |
0.1241563773 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 20 |
Hb_159295_010 |
0.1254232395 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |