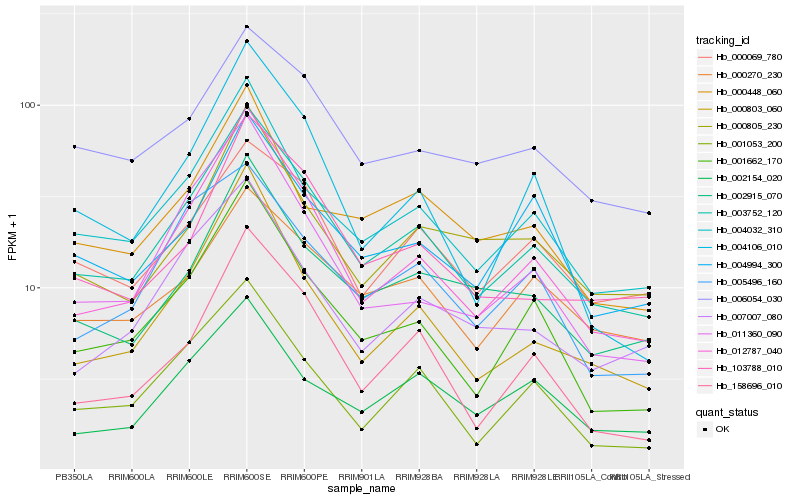
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003752_120 |
0.0 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 2 |
Hb_012787_040 |
0.0722961362 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004032_310 |
0.0724352966 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 4 |
Hb_001662_170 |
0.0939032853 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 5 |
Hb_002915_070 |
0.0983068915 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 6 |
Hb_000069_780 |
0.1001657297 |
- |
- |
atpob1, putative [Ricinus communis] |
| 7 |
Hb_103788_010 |
0.1001978533 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_006054_030 |
0.1032128822 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 9 |
Hb_000270_230 |
0.1053483137 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 10 |
Hb_000803_060 |
0.1072814035 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 11 |
Hb_002154_020 |
0.1075407103 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_000805_230 |
0.1078246509 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 13 |
Hb_007007_080 |
0.1082681796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004994_300 |
0.1104876844 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 15 |
Hb_001053_200 |
0.1106958329 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 16 |
Hb_000448_060 |
0.1117117595 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 17 |
Hb_158696_010 |
0.111712583 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 18 |
Hb_005496_160 |
0.1163856455 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011360_090 |
0.1185499108 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 20 |
Hb_004106_010 |
0.1214124055 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |