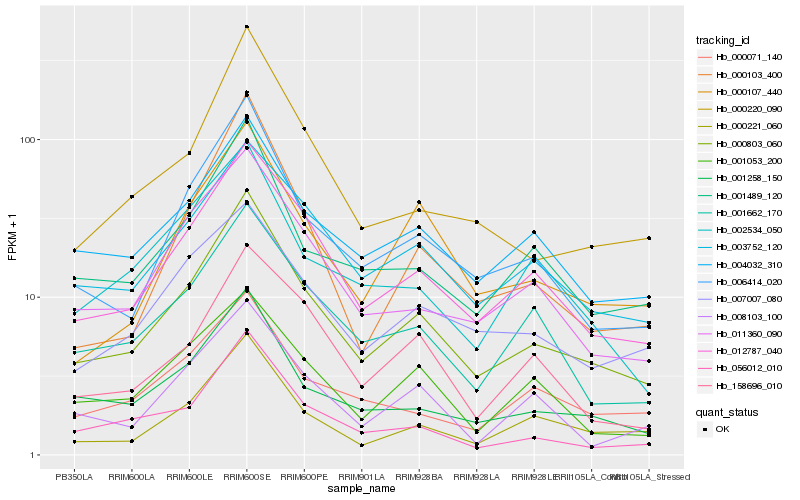
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_060 |
0.0 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 2 |
Hb_012787_040 |
0.0745127846 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 3 |
Hb_006414_020 |
0.0883318301 |
transcription factor |
TF Family: MYB |
- |
| 4 |
Hb_001258_150 |
0.1037282225 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 5 |
Hb_001489_120 |
0.1060484471 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003752_120 |
0.1072814035 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004032_310 |
0.1086713462 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 8 |
Hb_011360_090 |
0.1093442174 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 9 |
Hb_056012_010 |
0.1109996873 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 10 |
Hb_000221_060 |
0.1132831908 |
- |
- |
- |
| 11 |
Hb_001662_170 |
0.1136698807 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 12 |
Hb_000220_090 |
0.118452898 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 13 |
Hb_000107_440 |
0.1223220079 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 14 |
Hb_007007_080 |
0.123006521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008103_100 |
0.1279908477 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 16 |
Hb_000071_140 |
0.1294730619 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 17 |
Hb_001053_200 |
0.1302815944 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 18 |
Hb_158696_010 |
0.1309732276 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 19 |
Hb_000103_400 |
0.1311990702 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002534_050 |
0.1376152413 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |