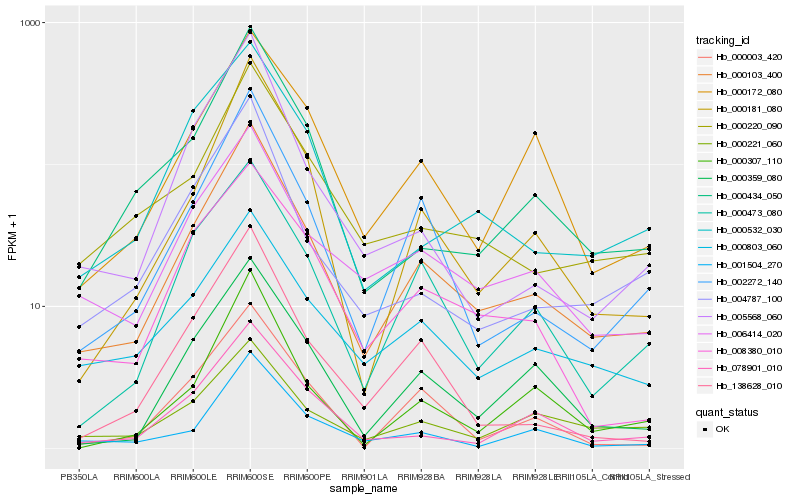
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_400 |
0.0 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002272_140 |
0.1136826283 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 3 |
Hb_000181_080 |
0.1167862878 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 4 |
Hb_000473_080 |
0.119713985 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 5 |
Hb_006414_020 |
0.1203359914 |
transcription factor |
TF Family: MYB |
- |
| 6 |
Hb_000359_080 |
0.1228004351 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 7 |
Hb_000803_060 |
0.1311990702 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 8 |
Hb_000434_050 |
0.1331577274 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 9 |
Hb_000220_090 |
0.1338563425 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 10 |
Hb_000003_420 |
0.1383851675 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_078901_010 |
0.1394160012 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 12 |
Hb_000172_080 |
0.139815375 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 13 |
Hb_008380_010 |
0.1399377381 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 14 |
Hb_004787_100 |
0.1407595533 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 15 |
Hb_000532_030 |
0.1420419509 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 16 |
Hb_001504_270 |
0.1423934801 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 17 |
Hb_000221_060 |
0.1451554031 |
- |
- |
- |
| 18 |
Hb_000307_110 |
0.1455625754 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 19 |
Hb_138628_010 |
0.1491331775 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_005568_060 |
0.1493450676 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |