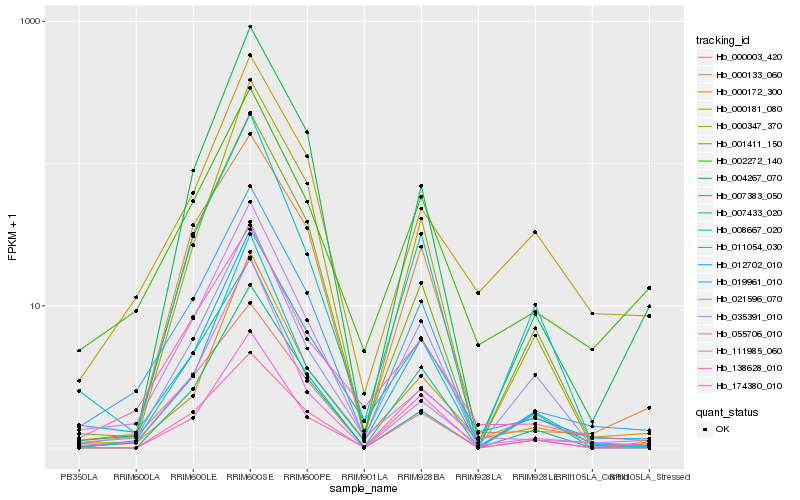
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019961_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 2 |
Hb_012702_010 |
0.098638637 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 3 |
Hb_138628_010 |
0.1111401882 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 4 |
Hb_000133_060 |
0.1149335098 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 5 |
Hb_002272_140 |
0.1158165239 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 6 |
Hb_007433_020 |
0.1180161831 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 7 |
Hb_008667_020 |
0.1295159225 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_000003_420 |
0.131311101 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_004267_070 |
0.134980422 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 10 |
Hb_021596_070 |
0.135997364 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 11 |
Hb_001411_150 |
0.1401391152 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 12 |
Hb_011054_030 |
0.1419124825 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 13 |
Hb_174380_010 |
0.1440686155 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000172_300 |
0.1443875624 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 15 |
Hb_000347_370 |
0.1449074215 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 16 |
Hb_000181_080 |
0.1477472825 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 17 |
Hb_111985_060 |
0.1485880731 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 18 |
Hb_055706_010 |
0.1498662356 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 19 |
Hb_007383_050 |
0.1520188628 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 20 |
Hb_035391_010 |
0.1548190487 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |