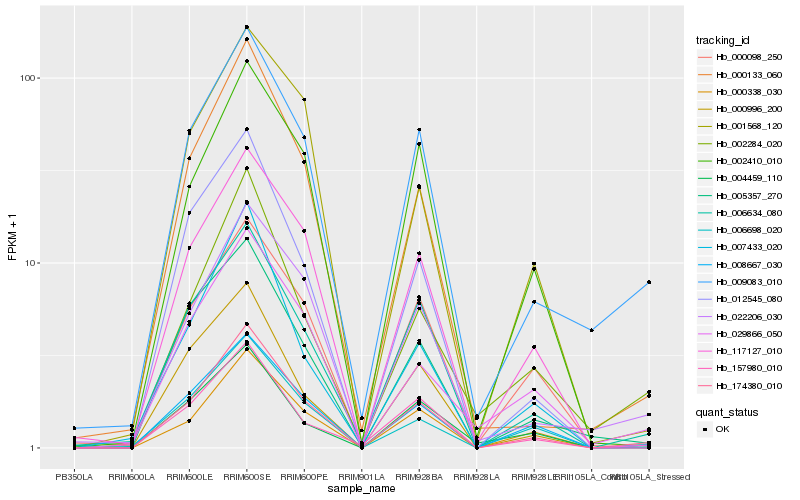
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174380_010 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_009083_010 |
0.1027437967 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 3 |
Hb_006634_080 |
0.1056865259 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 4 |
Hb_117127_010 |
0.1076538244 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 5 |
Hb_000338_030 |
0.1122270184 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |
| 6 |
Hb_000133_060 |
0.1161277981 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 7 |
Hb_012545_080 |
0.1190677334 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 8 |
Hb_000996_200 |
0.1198687319 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Populus euphratica] |
| 9 |
Hb_006698_020 |
0.1199957303 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 10 |
Hb_022206_030 |
0.121111167 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 11 |
Hb_008667_030 |
0.1215095331 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 12 |
Hb_002410_010 |
0.1222183366 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 13 |
Hb_005357_270 |
0.1241773752 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_007433_020 |
0.1264573388 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 15 |
Hb_002284_020 |
0.1279687441 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 16 |
Hb_029866_050 |
0.1291888732 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 17 |
Hb_157980_010 |
0.1380783978 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 18 |
Hb_000098_250 |
0.1381622791 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 19 |
Hb_001568_120 |
0.1383577054 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 20 |
Hb_004459_110 |
0.1388576781 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |