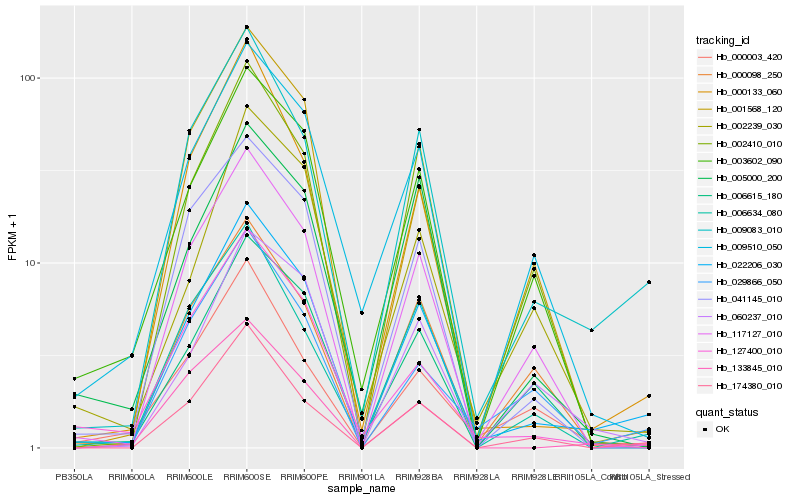
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022206_030 |
0.0 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 2 |
Hb_009083_010 |
0.081040164 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 3 |
Hb_174380_010 |
0.121111167 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_117127_010 |
0.1237385537 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 5 |
Hb_000133_060 |
0.1280887099 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 6 |
Hb_041145_010 |
0.1360164255 |
- |
- |
hypothetical protein CISIN_1g021653mg [Citrus sinensis] |
| 7 |
Hb_009510_050 |
0.1363760146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006634_080 |
0.1386633431 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 9 |
Hb_003602_090 |
0.139618411 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 10 |
Hb_002410_010 |
0.1425628532 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 11 |
Hb_060237_010 |
0.1427580689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_133845_010 |
0.1451224148 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_029866_050 |
0.1471849373 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 14 |
Hb_001568_120 |
0.1472971874 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 15 |
Hb_127400_010 |
0.1501291389 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_006615_180 |
0.1524370556 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 17 |
Hb_002239_030 |
0.1524958662 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 18 |
Hb_005000_200 |
0.1537354613 |
- |
- |
PREDICTED: uncharacterized protein LOC105637887 [Jatropha curcas] |
| 19 |
Hb_000003_420 |
0.154004793 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000098_250 |
0.1552666027 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |