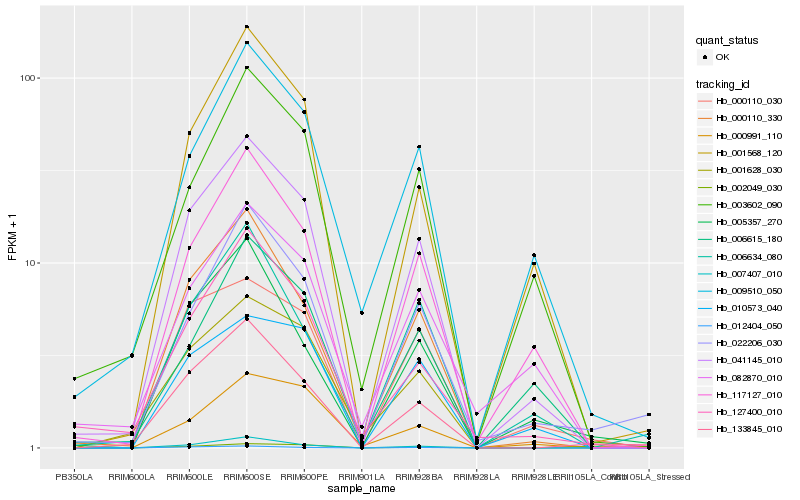
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041145_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g021653mg [Citrus sinensis] |
| 2 |
Hb_000110_330 |
0.090401896 |
- |
- |
PREDICTED: probable nitrite transporter At1g68570-like [Glycine max] |
| 3 |
Hb_117127_010 |
0.1013935337 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 4 |
Hb_001568_120 |
0.1200577452 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 5 |
Hb_133845_010 |
0.1268238784 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_003602_090 |
0.1293308407 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 7 |
Hb_127400_010 |
0.1313642465 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_009510_050 |
0.1314656525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001628_030 |
0.1324640352 |
- |
- |
PREDICTED: uncharacterized protein LOC105629266 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000110_030 |
0.133298287 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_007407_010 |
0.1338156991 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 74 [Populus euphratica] |
| 12 |
Hb_010573_040 |
0.1355146906 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 13 |
Hb_022206_030 |
0.1360164255 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 14 |
Hb_012404_050 |
0.1393201987 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 15 |
Hb_006634_080 |
0.14106004 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 16 |
Hb_002049_030 |
0.1420273699 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |
| 17 |
Hb_005357_270 |
0.142423213 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_082870_010 |
0.1434549654 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 19 |
Hb_000991_110 |
0.143973485 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 20 |
Hb_006615_180 |
0.144552063 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |