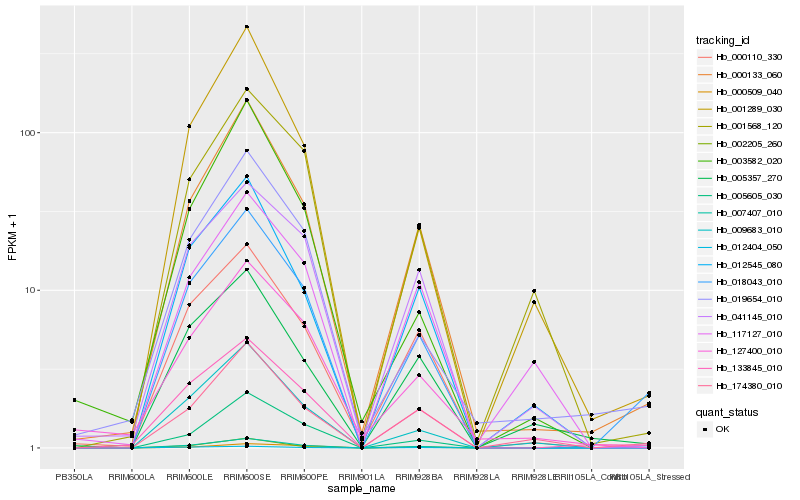
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007407_010 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 74 [Populus euphratica] |
| 2 |
Hb_000133_060 |
0.0771566368 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 3 |
Hb_005605_030 |
0.0919577348 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 4 |
Hb_000110_330 |
0.0998569704 |
- |
- |
PREDICTED: probable nitrite transporter At1g68570-like [Glycine max] |
| 5 |
Hb_002205_260 |
0.1023928026 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 6 |
Hb_009683_010 |
0.1067877802 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_012545_080 |
0.1090348721 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 8 |
Hb_018043_010 |
0.1117711884 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 9 |
Hb_133845_010 |
0.1150165455 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_012404_050 |
0.1229181551 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 11 |
Hb_041145_010 |
0.1338156991 |
- |
- |
hypothetical protein CISIN_1g021653mg [Citrus sinensis] |
| 12 |
Hb_001289_030 |
0.1384009993 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 13 |
Hb_001568_120 |
0.1404145867 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 14 |
Hb_174380_010 |
0.1434380384 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_003582_020 |
0.1451977139 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 16 |
Hb_000509_040 |
0.1485072482 |
- |
- |
hypothetical protein VITISV_009716 [Vitis vinifera] |
| 17 |
Hb_005357_270 |
0.1497141431 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_127400_010 |
0.1506641464 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 19 |
Hb_019654_010 |
0.1527694095 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 20 |
Hb_117127_010 |
0.1539560468 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |