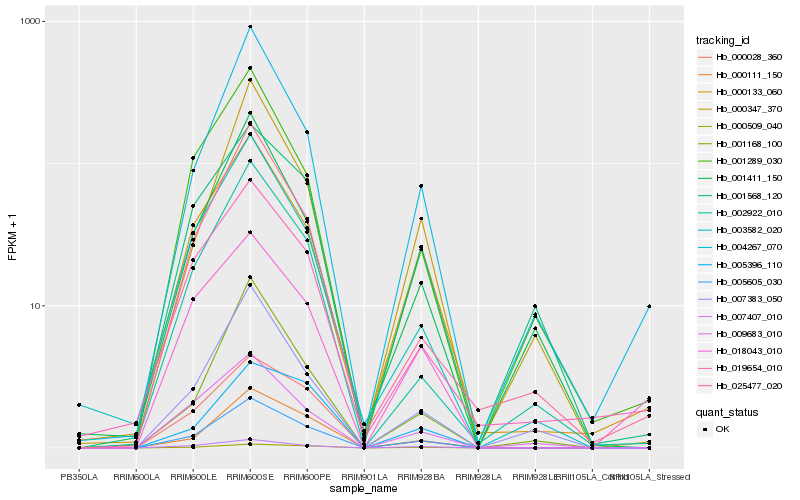
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005605_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 2 |
Hb_000111_150 |
0.0901377674 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 3 |
Hb_007407_010 |
0.0919577348 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 74 [Populus euphratica] |
| 4 |
Hb_000509_040 |
0.0968646784 |
- |
- |
hypothetical protein VITISV_009716 [Vitis vinifera] |
| 5 |
Hb_000028_360 |
0.1072343412 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000133_060 |
0.1225612362 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 7 |
Hb_002922_010 |
0.1288197638 |
- |
- |
heavy-metal-associated domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_003582_020 |
0.130740908 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 9 |
Hb_005396_110 |
0.1307756259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001568_120 |
0.1342477931 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 11 |
Hb_009683_010 |
0.1352925635 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_004267_070 |
0.1376358776 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 13 |
Hb_019654_010 |
0.1414696418 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 14 |
Hb_007383_050 |
0.1417792432 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 15 |
Hb_025477_020 |
0.1421213005 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 16 |
Hb_018043_010 |
0.1438997944 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 17 |
Hb_001168_100 |
0.1448511287 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 18 |
Hb_001289_030 |
0.1481965855 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 19 |
Hb_001411_150 |
0.1497780606 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 20 |
Hb_000347_370 |
0.1508226687 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |