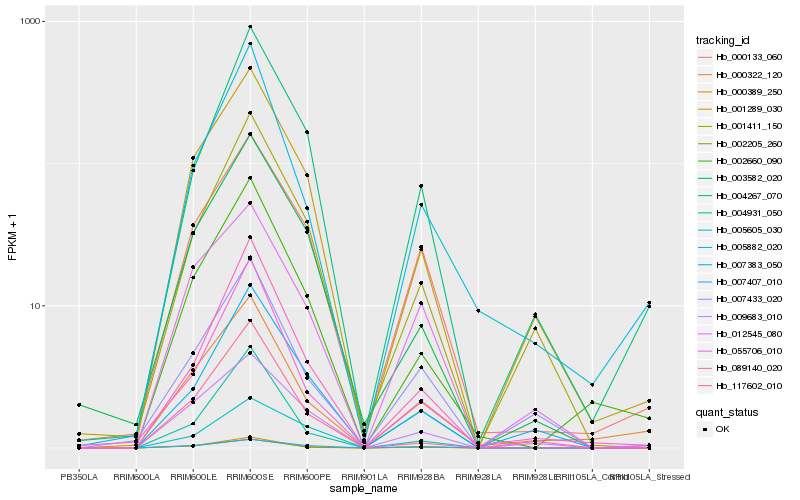
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_260 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 2 |
Hb_007407_010 |
0.1023928026 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 74 [Populus euphratica] |
| 3 |
Hb_000133_060 |
0.1065593192 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 4 |
Hb_012545_080 |
0.1160778659 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 5 |
Hb_001289_030 |
0.116212274 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 6 |
Hb_002660_090 |
0.1217319942 |
- |
- |
hypothetical protein JCGZ_16500 [Jatropha curcas] |
| 7 |
Hb_089140_020 |
0.1251840892 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_009683_010 |
0.125662276 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_003582_020 |
0.1337841992 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 10 |
Hb_007433_020 |
0.1345718434 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 11 |
Hb_001411_150 |
0.1408045491 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 12 |
Hb_000322_120 |
0.1416511368 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 13 |
Hb_004931_050 |
0.1416622534 |
transcription factor |
TF Family: MYB-related |
Transcription repressor MYB4, putative [Ricinus communis] |
| 14 |
Hb_007383_050 |
0.1440614819 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 15 |
Hb_005882_020 |
0.1459378377 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 16 |
Hb_117602_010 |
0.1460161558 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 17 |
Hb_000389_250 |
0.1499239181 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 18 |
Hb_055706_010 |
0.1506887395 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 19 |
Hb_004267_070 |
0.1516183656 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 20 |
Hb_005605_030 |
0.1519692456 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |