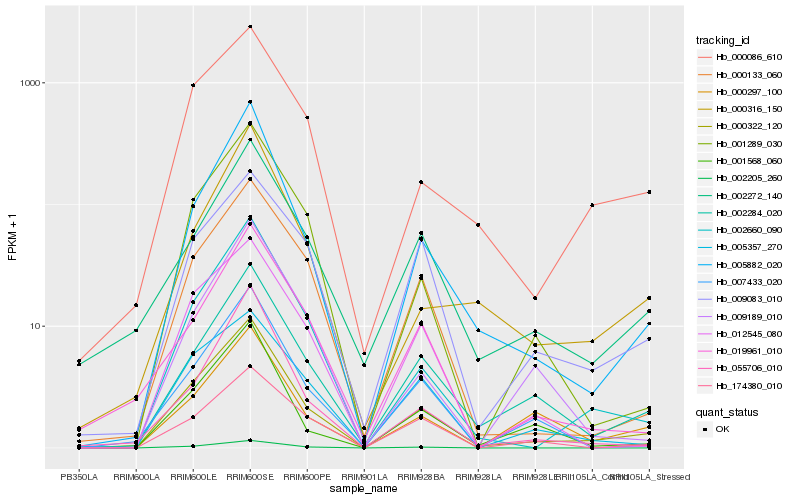
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000322_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 2 |
Hb_005882_020 |
0.1102525593 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 3 |
Hb_002660_090 |
0.1190603066 |
- |
- |
hypothetical protein JCGZ_16500 [Jatropha curcas] |
| 4 |
Hb_002284_020 |
0.1275318821 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 5 |
Hb_000086_610 |
0.1383469292 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 6 |
Hb_001289_030 |
0.139967749 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 7 |
Hb_002205_260 |
0.1416511368 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 8 |
Hb_000133_060 |
0.1424294873 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_001568_060 |
0.1438158991 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 10 |
Hb_007433_020 |
0.1445774482 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 11 |
Hb_012545_080 |
0.1519850846 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 12 |
Hb_055706_010 |
0.15389877 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 13 |
Hb_174380_010 |
0.1549485224 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_005357_270 |
0.1599350665 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000316_150 |
0.1608024127 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_000297_100 |
0.1609596083 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 17 |
Hb_009083_010 |
0.1609995828 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 18 |
Hb_019961_010 |
0.1614337641 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 19 |
Hb_002272_140 |
0.1627418219 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 20 |
Hb_009189_010 |
0.1646905275 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |