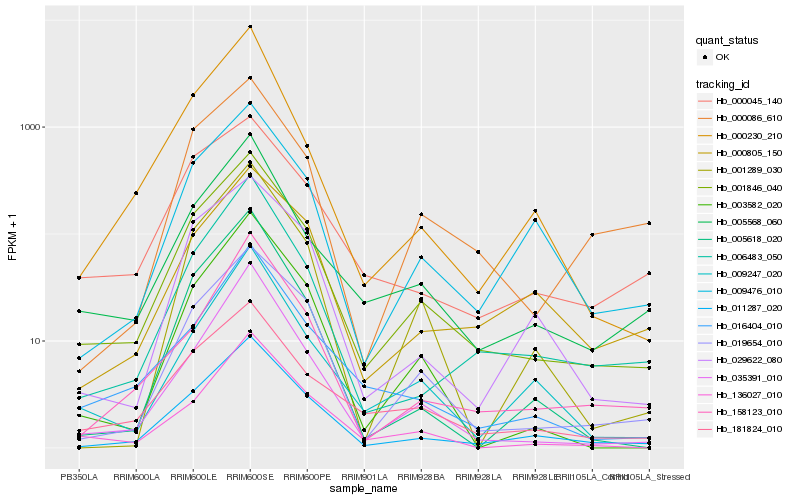
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001846_040 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_011287_020 |
0.079728028 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_019654_010 |
0.0994809648 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 4 |
Hb_005568_060 |
0.1038700558 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 5 |
Hb_181824_010 |
0.1077187556 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_006483_050 |
0.1182811377 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 7 |
Hb_035391_010 |
0.1204044217 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 8 |
Hb_016404_010 |
0.1206999738 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 9 |
Hb_136027_010 |
0.1229085281 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_003582_020 |
0.1229416009 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 11 |
Hb_000045_140 |
0.1259357963 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 12 |
Hb_009476_010 |
0.128488056 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 13 |
Hb_000086_610 |
0.1297043595 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 14 |
Hb_158123_010 |
0.1335044628 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 15 |
Hb_029622_080 |
0.1352016921 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 16 |
Hb_001289_030 |
0.1364185156 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 17 |
Hb_009247_020 |
0.1377180697 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 18 |
Hb_000230_210 |
0.1431811891 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 19 |
Hb_005618_020 |
0.1440937293 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 20 |
Hb_000805_150 |
0.1448465376 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |