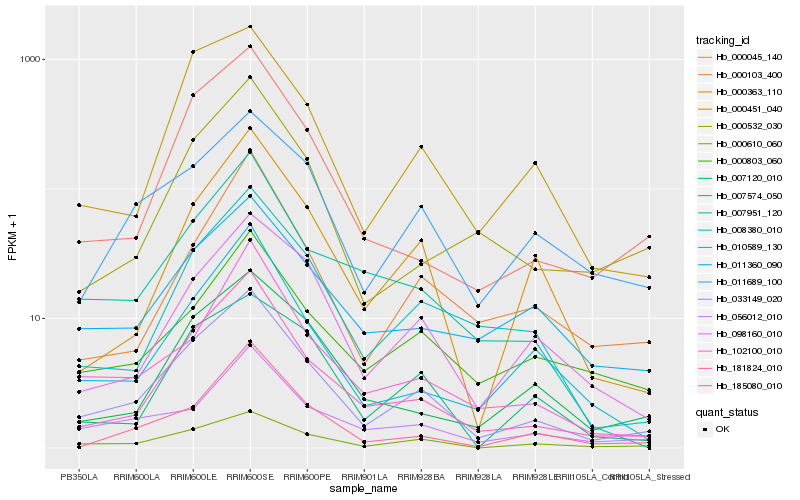
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033149_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 2 |
Hb_000610_060 |
0.1027322137 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 3 |
Hb_181824_010 |
0.1057443529 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_000451_040 |
0.1324675127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008380_010 |
0.1403903556 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 6 |
Hb_000045_140 |
0.1430026315 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 7 |
Hb_056012_010 |
0.1442218308 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 8 |
Hb_098160_010 |
0.1461543362 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_102100_010 |
0.1498177276 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 10 |
Hb_011689_100 |
0.1509237401 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 11 |
Hb_007951_120 |
0.1527277453 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 12 |
Hb_000803_060 |
0.1535557587 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 13 |
Hb_000363_110 |
0.1543264551 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 14 |
Hb_007120_010 |
0.1548015267 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 15 |
Hb_011360_090 |
0.1548283136 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 16 |
Hb_010589_130 |
0.1554954302 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_007574_050 |
0.1589763248 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 18 |
Hb_000532_030 |
0.1592546624 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 19 |
Hb_185080_010 |
0.1593678523 |
- |
- |
- |
| 20 |
Hb_000103_400 |
0.1599329901 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |