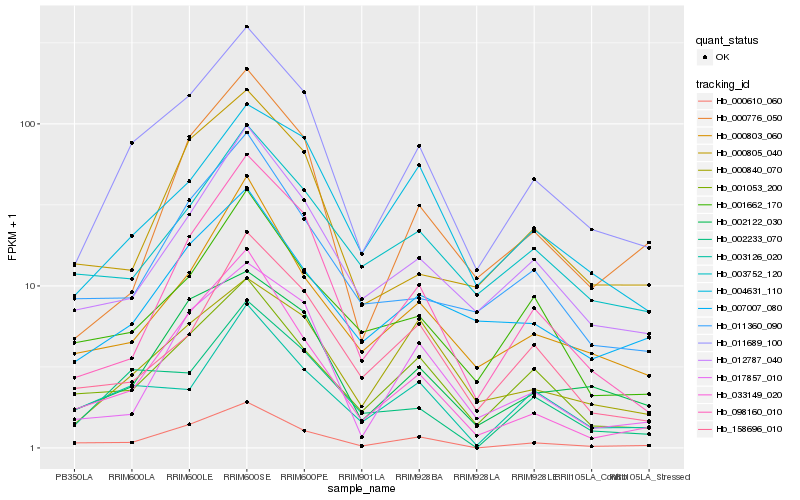
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011689_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 2 |
Hb_002122_030 |
0.1231184605 |
- |
- |
- |
| 3 |
Hb_012787_040 |
0.1274599394 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001053_200 |
0.1362745463 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 5 |
Hb_000610_060 |
0.139433407 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 6 |
Hb_000803_060 |
0.1402546598 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 7 |
Hb_002233_070 |
0.1419830804 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_098160_010 |
0.1431999791 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000840_070 |
0.1467799092 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000776_050 |
0.1481730452 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 11 |
Hb_007007_080 |
0.1481874568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004631_110 |
0.1498517147 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_011360_090 |
0.1504141967 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 14 |
Hb_033149_020 |
0.1509237401 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 15 |
Hb_017857_010 |
0.1534537777 |
- |
- |
- |
| 16 |
Hb_001662_170 |
0.1536173617 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 17 |
Hb_000805_040 |
0.1537638282 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 18 |
Hb_003752_120 |
0.1547987758 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 19 |
Hb_158696_010 |
0.156854003 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 20 |
Hb_003126_020 |
0.1578729836 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |