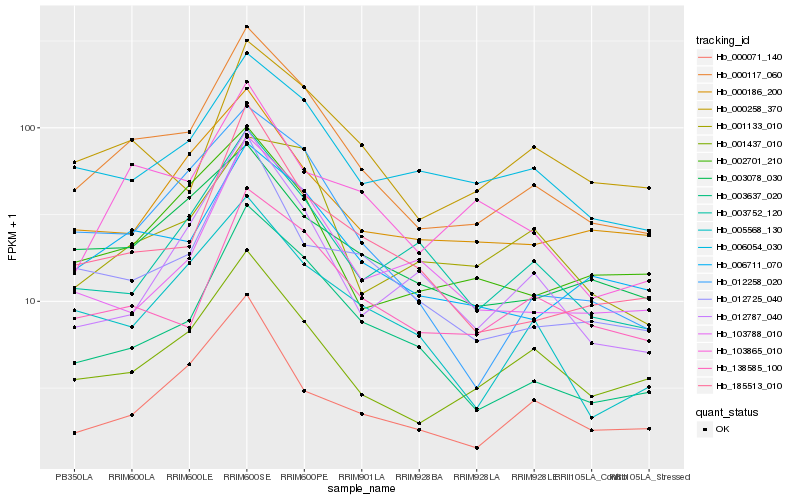
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000117_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 2 |
Hb_006711_070 |
0.1057246086 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 3 |
Hb_003637_020 |
0.1111343473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_185513_010 |
0.1249526547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001437_010 |
0.1285430506 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 6 |
Hb_012258_020 |
0.1306430216 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_138585_100 |
0.1339038758 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 8 |
Hb_006054_030 |
0.1360921061 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 9 |
Hb_012725_040 |
0.1366461814 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_005568_130 |
0.1402337909 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_002701_210 |
0.1414828045 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 12 |
Hb_103865_010 |
0.1422394925 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 13 |
Hb_103788_010 |
0.1428500803 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 14 |
Hb_003752_120 |
0.1428509758 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000258_370 |
0.1441350022 |
- |
- |
hypothetical protein [Ricinus communis] |
| 16 |
Hb_003078_030 |
0.1446746595 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 17 |
Hb_000186_200 |
0.1468930829 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 18 |
Hb_012787_040 |
0.1500574182 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000071_140 |
0.1508176776 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 20 |
Hb_001133_010 |
0.1519456166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |