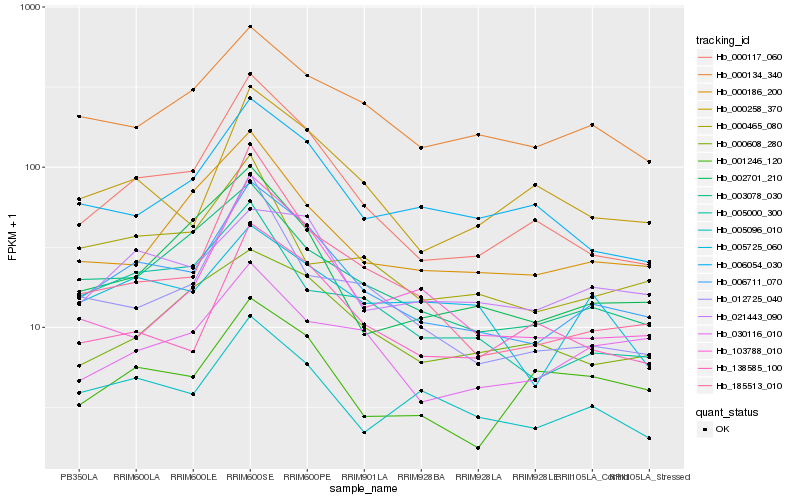
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006711_070 |
0.0 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 2 |
Hb_000117_060 |
0.1057246086 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 3 |
Hb_003078_030 |
0.1162273289 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 4 |
Hb_000134_340 |
0.1220437417 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 5 |
Hb_005096_010 |
0.1252637949 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 6 |
Hb_138585_100 |
0.1265787917 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 7 |
Hb_021443_090 |
0.1277522159 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 8 |
Hb_000258_370 |
0.1278075735 |
- |
- |
hypothetical protein [Ricinus communis] |
| 9 |
Hb_006054_030 |
0.1336545441 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 10 |
Hb_002701_210 |
0.134761989 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 11 |
Hb_005000_300 |
0.1355885998 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 12 |
Hb_000186_200 |
0.135746524 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 13 |
Hb_000608_280 |
0.1385913816 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 14 |
Hb_030116_010 |
0.1422059515 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 15 |
Hb_185513_010 |
0.1429669262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_103788_010 |
0.1470416076 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_000465_080 |
0.1479625674 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 18 |
Hb_005725_060 |
0.1489842926 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 19 |
Hb_012725_040 |
0.1494996201 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_001246_120 |
0.1495690648 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |