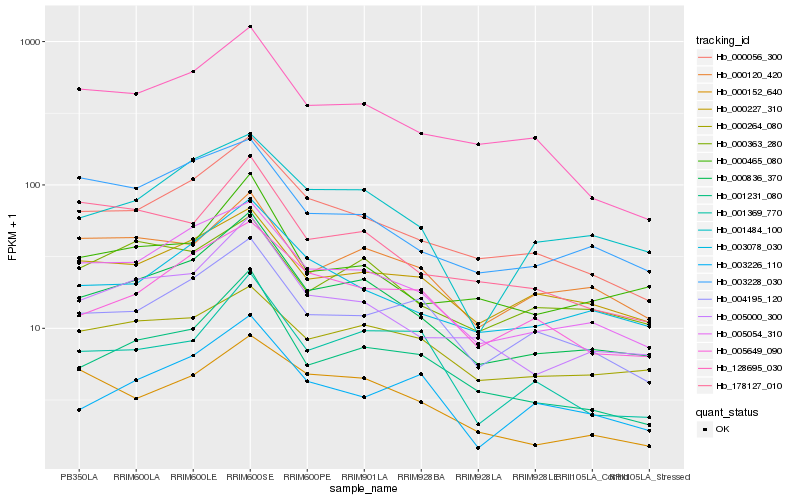
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_370 |
0.0 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_005054_310 |
0.0696834327 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 3 |
Hb_005000_300 |
0.072644327 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 4 |
Hb_000056_300 |
0.0813356373 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001231_080 |
0.0860480105 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_003078_030 |
0.0958352855 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 7 |
Hb_001484_100 |
0.1016873771 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 8 |
Hb_128695_030 |
0.1030196517 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 9 |
Hb_000465_080 |
0.1081047559 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 10 |
Hb_005649_090 |
0.1101114897 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 11 |
Hb_004195_120 |
0.1174813683 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 12 |
Hb_003228_030 |
0.1192151831 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_000227_310 |
0.120849962 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000120_420 |
0.1210895767 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 15 |
Hb_178127_010 |
0.1214681671 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 16 |
Hb_001369_770 |
0.1262577702 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 17 |
Hb_000363_280 |
0.127012923 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 18 |
Hb_000152_640 |
0.1280514163 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 19 |
Hb_000264_080 |
0.1305236302 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 20 |
Hb_003226_110 |
0.1318405006 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |