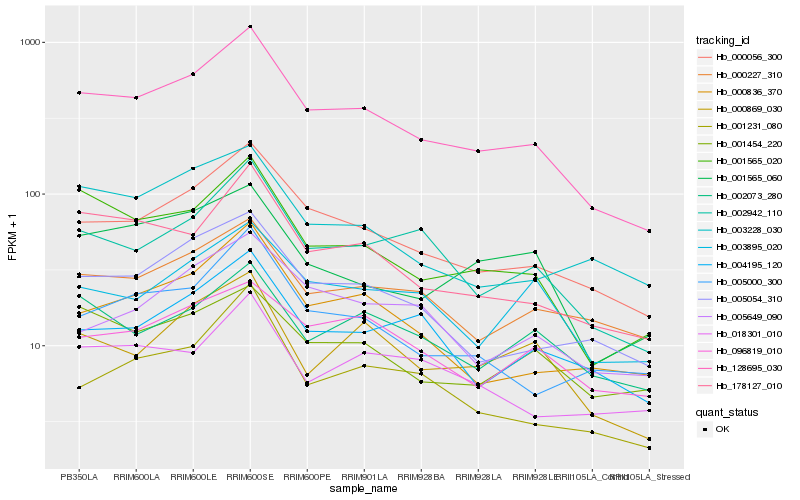
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_128695_030 |
0.0 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 2 |
Hb_000056_300 |
0.0646992901 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_178127_010 |
0.0882903844 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 4 |
Hb_001565_020 |
0.089768533 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |
| 5 |
Hb_002942_110 |
0.094605394 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 6 |
Hb_005054_310 |
0.0950706939 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 7 |
Hb_000836_370 |
0.1030196517 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005000_300 |
0.1101089638 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 9 |
Hb_001231_080 |
0.1117407138 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000869_030 |
0.1139078536 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000227_310 |
0.1163210372 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_004195_120 |
0.1165796562 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 13 |
Hb_003228_030 |
0.1170443766 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_001565_060 |
0.1239235779 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001454_220 |
0.124080203 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 16 |
Hb_003895_020 |
0.1249648487 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 17 |
Hb_005649_090 |
0.1260123524 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 18 |
Hb_096819_010 |
0.1261754792 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 19 |
Hb_002073_280 |
0.126432129 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_018301_010 |
0.1264573221 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |