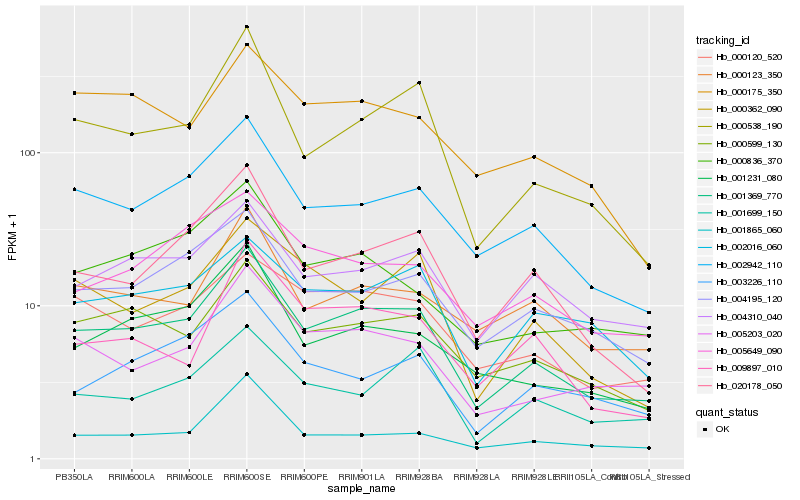
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_770 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 2 |
Hb_002942_110 |
0.0953222944 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 3 |
Hb_000538_190 |
0.1013800714 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 4 |
Hb_000599_130 |
0.1016876136 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 5 |
Hb_020178_050 |
0.1030472491 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 6 |
Hb_004195_120 |
0.1105522406 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_005203_020 |
0.1110056464 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 8 |
Hb_001231_080 |
0.1149763701 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_000120_520 |
0.1180626271 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 10 |
Hb_004310_040 |
0.1186860727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000123_350 |
0.1255012491 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000836_370 |
0.1262577702 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002016_060 |
0.127264086 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 14 |
Hb_005649_090 |
0.1295960405 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 15 |
Hb_000362_090 |
0.1297658721 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003226_110 |
0.1318673322 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 17 |
Hb_000175_350 |
0.1327510117 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 18 |
Hb_001699_150 |
0.1331539048 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 19 |
Hb_001865_060 |
0.1336371239 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 20 |
Hb_009897_010 |
0.1343335597 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |