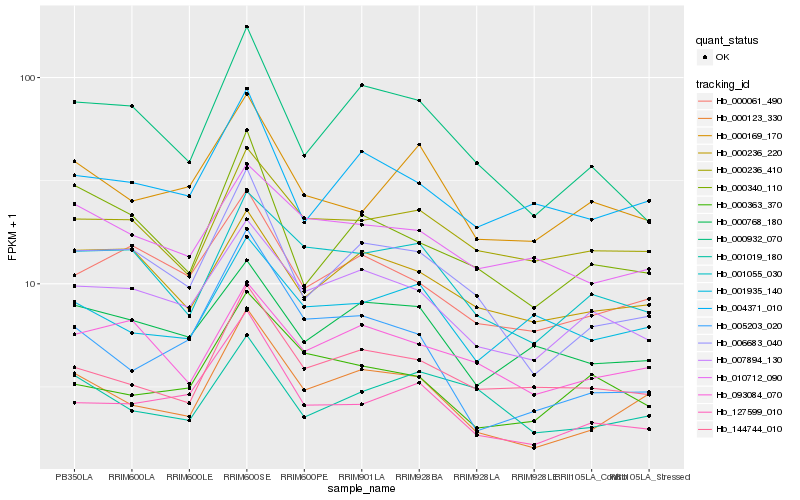
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000123_330 |
0.0 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 2 |
Hb_006683_040 |
0.1075060348 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_144744_010 |
0.1150364584 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 4 |
Hb_007894_130 |
0.1164764498 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 5 |
Hb_001055_030 |
0.1173558183 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 6 |
Hb_127599_010 |
0.1212132402 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 7 |
Hb_005203_020 |
0.1233746559 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 8 |
Hb_004371_010 |
0.1255453408 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_000061_490 |
0.1278761742 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 10 |
Hb_000236_410 |
0.1285619019 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 11 |
Hb_000340_110 |
0.1300136159 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 12 |
Hb_000932_070 |
0.1312063781 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 13 |
Hb_000363_370 |
0.131500141 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 14 |
Hb_000768_180 |
0.1333022037 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 15 |
Hb_001935_140 |
0.1333530113 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 16 |
Hb_093084_070 |
0.133564975 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 17 |
Hb_000169_170 |
0.1349002498 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 18 |
Hb_010712_090 |
0.1352780691 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 19 |
Hb_000236_220 |
0.1383936845 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 20 |
Hb_001019_180 |
0.1391355307 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |