| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
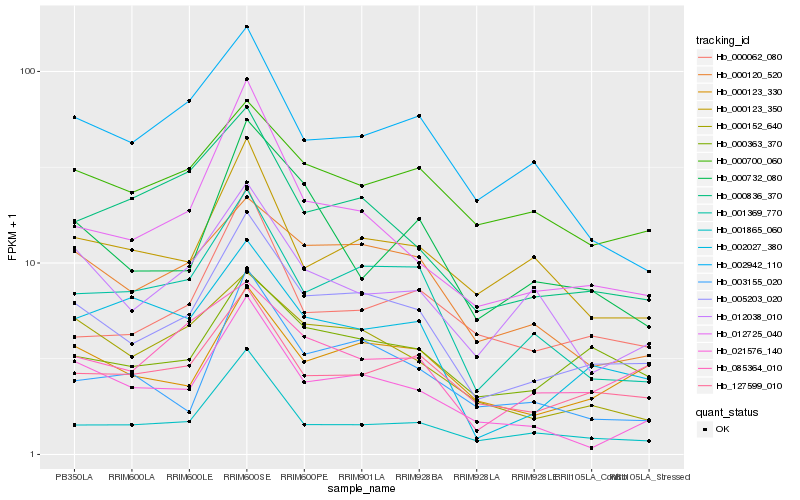
Hb_005203_020 |
0.0 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 2 |
Hb_001369_770 |
0.1110056464 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 3 |
Hb_127599_010 |
0.1163661995 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 4 |
Hb_000120_520 |
0.1214847349 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 5 |
Hb_000123_330 |
0.1233746559 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 6 |
Hb_000363_370 |
0.1240305839 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 7 |
Hb_012725_040 |
0.1243395504 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000152_640 |
0.1271162388 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 9 |
Hb_021576_140 |
0.1296663281 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 10 |
Hb_001865_060 |
0.1313167542 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 11 |
Hb_085364_010 |
0.1344937682 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 12 |
Hb_000836_370 |
0.136255892 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_012038_010 |
0.1395046008 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 14 |
Hb_000123_350 |
0.1430624529 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_003155_020 |
0.1437977536 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 16 |
Hb_002027_380 |
0.1439765464 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 17 |
Hb_000732_080 |
0.1442115435 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |
| 18 |
Hb_000700_060 |
0.1454363748 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 19 |
Hb_000062_080 |
0.145849294 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 20 |
Hb_002942_110 |
0.1459780897 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |