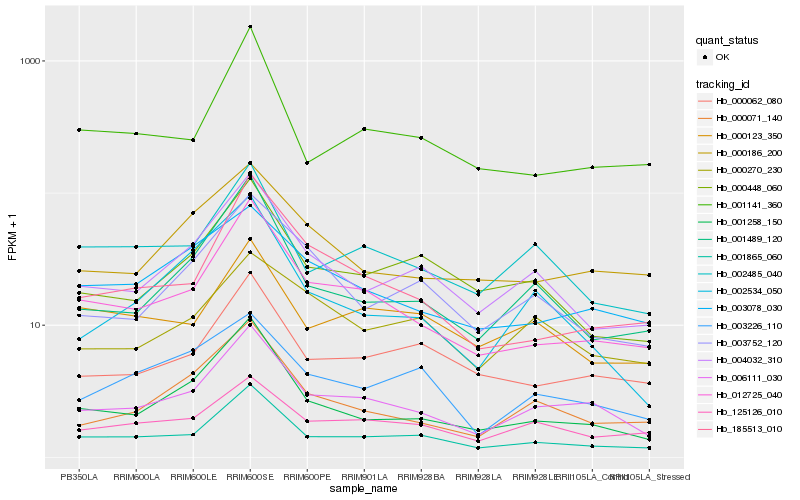
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006111_030 |
0.0 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001865_060 |
0.1009393788 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 3 |
Hb_012725_040 |
0.109320568 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000071_140 |
0.1260141941 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 5 |
Hb_001258_150 |
0.1264133211 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 6 |
Hb_004032_310 |
0.1268041148 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_002485_040 |
0.1278297899 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 8 |
Hb_125126_010 |
0.1333814892 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 9 |
Hb_000062_080 |
0.1341211871 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 10 |
Hb_001489_120 |
0.1352408815 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003078_030 |
0.1364460612 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 12 |
Hb_000186_200 |
0.1386368555 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 13 |
Hb_000448_060 |
0.1468012242 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 14 |
Hb_003752_120 |
0.1483289714 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003226_110 |
0.1483295708 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 16 |
Hb_185513_010 |
0.148538997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000270_230 |
0.1487226278 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 18 |
Hb_001141_360 |
0.1493513989 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 19 |
Hb_002534_050 |
0.1519122134 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 20 |
Hb_000123_350 |
0.1521677334 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |