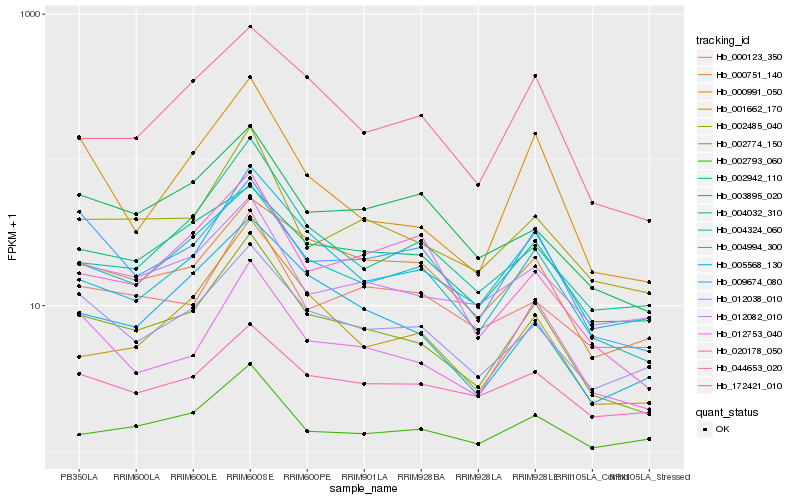
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002774_150 |
0.0 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 2 |
Hb_004324_060 |
0.0844498204 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 3 |
Hb_002485_040 |
0.108388361 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 4 |
Hb_012038_010 |
0.1202135001 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 5 |
Hb_172421_010 |
0.1216656871 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 6 |
Hb_004994_300 |
0.1228905963 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 7 |
Hb_012753_040 |
0.1230255175 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005568_130 |
0.126463766 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_004032_310 |
0.1296921366 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 10 |
Hb_009674_080 |
0.1309049645 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 11 |
Hb_001662_170 |
0.1332441986 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 12 |
Hb_003895_020 |
0.1339935546 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000751_140 |
0.1350121406 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 14 |
Hb_002942_110 |
0.1353224776 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 15 |
Hb_000991_050 |
0.1354850383 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 16 |
Hb_012082_010 |
0.1410022158 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000123_350 |
0.1430437504 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_002793_060 |
0.1440785143 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 19 |
Hb_020178_050 |
0.1441257362 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 20 |
Hb_044653_020 |
0.1446591996 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |