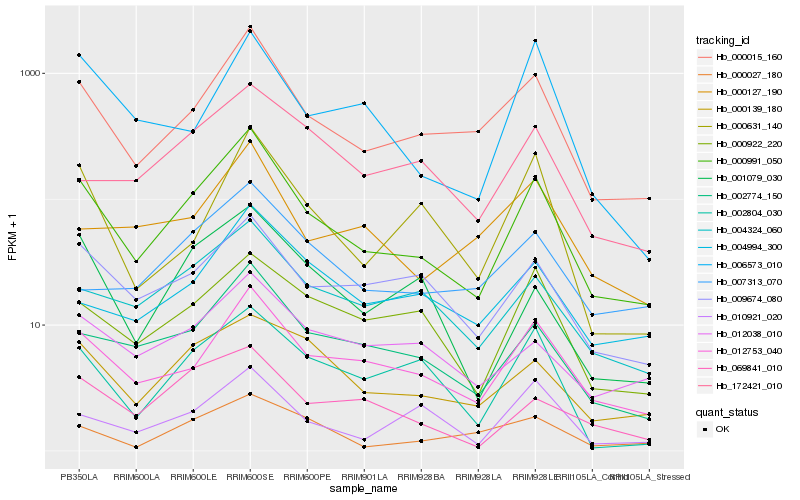
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000991_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 2 |
Hb_000015_160 |
0.1072312019 |
- |
- |
Heat shock cognate 70 kDa protein 1 [Triticum urartu] |
| 3 |
Hb_012753_040 |
0.1189340973 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002774_150 |
0.1354850383 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 5 |
Hb_009674_080 |
0.1573354857 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 6 |
Hb_004324_060 |
0.1582807588 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 7 |
Hb_000139_180 |
0.1583929469 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 8 |
Hb_000922_220 |
0.1662477827 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 9 |
Hb_069841_010 |
0.1681657903 |
- |
- |
resistance protein [Quercus suber] |
| 10 |
Hb_012038_010 |
0.1690935528 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 11 |
Hb_002804_030 |
0.1707213321 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 12 |
Hb_001079_030 |
0.1714162831 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 13 |
Hb_000631_140 |
0.1724441954 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 14 |
Hb_004994_300 |
0.1780660588 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 15 |
Hb_000027_180 |
0.1854661213 |
- |
- |
PREDICTED: uncharacterized protein LOC105642058 [Jatropha curcas] |
| 16 |
Hb_010921_020 |
0.1856295934 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 17 |
Hb_172421_010 |
0.1865694179 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 18 |
Hb_000127_190 |
0.1866588741 |
- |
- |
PREDICTED: protein GIGANTEA [Jatropha curcas] |
| 19 |
Hb_007313_070 |
0.1867914027 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 20 |
Hb_006573_010 |
0.1873870961 |
- |
- |
heat shock protein [Hevea brasiliensis] |