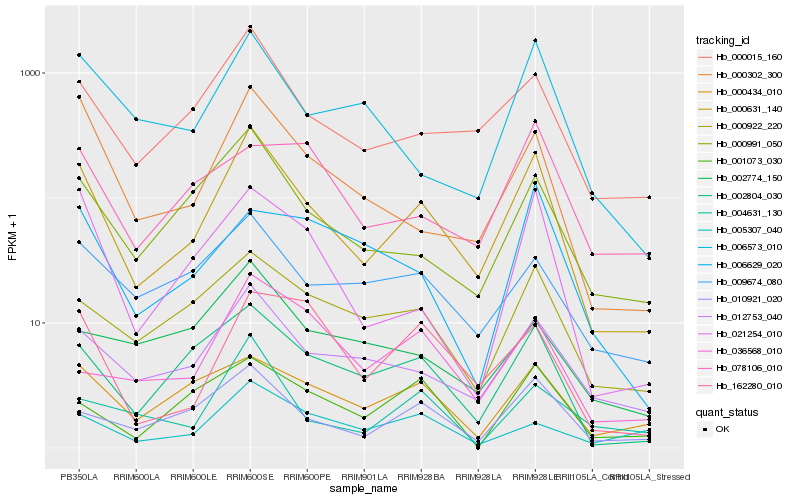
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000631_140 |
0.0 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 2 |
Hb_000015_160 |
0.1514714549 |
- |
- |
Heat shock cognate 70 kDa protein 1 [Triticum urartu] |
| 3 |
Hb_012753_040 |
0.1572272211 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002804_030 |
0.1644338895 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 5 |
Hb_010921_020 |
0.1650965539 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 6 |
Hb_000991_050 |
0.1724441954 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 7 |
Hb_000302_300 |
0.1737196068 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 8 |
Hb_021254_010 |
0.1822938225 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 9 |
Hb_000922_220 |
0.1883553655 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 10 |
Hb_006573_010 |
0.1959266603 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 11 |
Hb_000434_010 |
0.1962008358 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 12 |
Hb_009674_080 |
0.1987158662 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 13 |
Hb_162280_010 |
0.2084285578 |
- |
- |
Phosphoprotein phosphatase isoform 2, partial [Theobroma cacao] |
| 14 |
Hb_004631_130 |
0.219907581 |
- |
- |
helicase domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_036568_010 |
0.223416672 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 16 |
Hb_001073_030 |
0.227033165 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005307_040 |
0.227589239 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 18 |
Hb_002774_150 |
0.2299157681 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 19 |
Hb_078106_010 |
0.2300456695 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |
| 20 |
Hb_006629_020 |
0.2326119574 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |