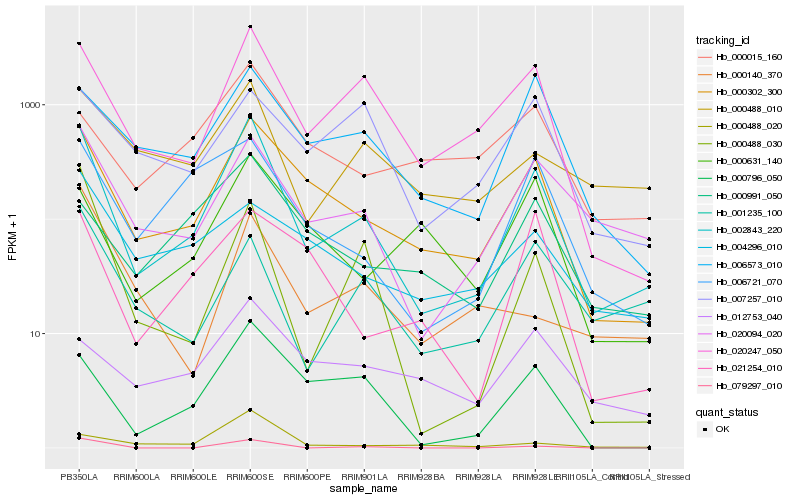
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002843_220 |
0.0 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000302_300 |
0.1581626795 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 3 |
Hb_020094_020 |
0.1844067857 |
- |
- |
PREDICTED: 17.1 kDa class II heat shock protein-like [Pyrus x bretschneideri] |
| 4 |
Hb_020247_050 |
0.1848005773 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 5 |
Hb_000796_050 |
0.2160584903 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 6 |
Hb_006721_070 |
0.2200607767 |
- |
- |
PREDICTED: 70 kDa peptidyl-prolyl isomerase-like [Jatropha curcas] |
| 7 |
Hb_006573_010 |
0.2285314162 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 8 |
Hb_000488_010 |
0.2359919453 |
- |
- |
18.1 kDa class I heat shock protein [Jatropha curcas] |
| 9 |
Hb_001235_100 |
0.2401773515 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 10 |
Hb_000488_030 |
0.2403575949 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 11 |
Hb_000488_020 |
0.2463104996 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 12 |
Hb_000991_050 |
0.2485847217 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 13 |
Hb_021254_010 |
0.2572882024 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 14 |
Hb_000631_140 |
0.2611340491 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 15 |
Hb_004296_010 |
0.2649325904 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 16 |
Hb_012753_040 |
0.266101416 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 17 |
Hb_079297_010 |
0.26644388 |
- |
- |
hypothetical protein VITISV_012409 [Vitis vinifera] |
| 18 |
Hb_000140_370 |
0.2678522723 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6 [Jatropha curcas] |
| 19 |
Hb_000015_160 |
0.2698689927 |
- |
- |
Heat shock cognate 70 kDa protein 1 [Triticum urartu] |
| 20 |
Hb_007257_010 |
0.2700809296 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |