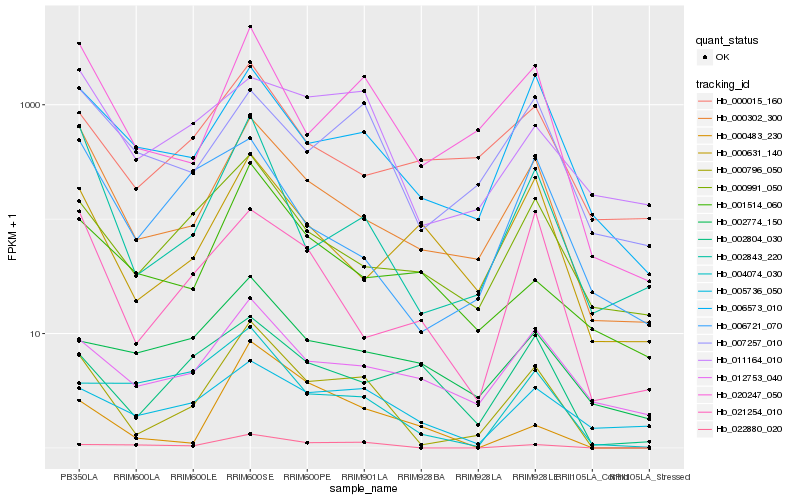
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000796_050 |
0.0 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 2 |
Hb_000302_300 |
0.1891546723 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 3 |
Hb_020247_050 |
0.19171452 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 4 |
Hb_002843_220 |
0.2160584903 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006573_010 |
0.2220845231 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 6 |
Hb_022880_020 |
0.2229954971 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 7 |
Hb_000991_050 |
0.2397856661 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 8 |
Hb_012753_040 |
0.2466559201 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007257_010 |
0.2508025828 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 10 |
Hb_021254_010 |
0.2599542741 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 11 |
Hb_004074_030 |
0.2613308012 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_011164_010 |
0.2620365084 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 13 |
Hb_000631_140 |
0.2665295362 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 14 |
Hb_000015_160 |
0.2683298557 |
- |
- |
Heat shock cognate 70 kDa protein 1 [Triticum urartu] |
| 15 |
Hb_005736_050 |
0.2692271865 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 16 |
Hb_002804_030 |
0.2740743834 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 17 |
Hb_001514_060 |
0.2753334615 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 18 |
Hb_006721_070 |
0.2760819911 |
- |
- |
PREDICTED: 70 kDa peptidyl-prolyl isomerase-like [Jatropha curcas] |
| 19 |
Hb_002774_150 |
0.2773712698 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 20 |
Hb_000483_230 |
0.2780215172 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |