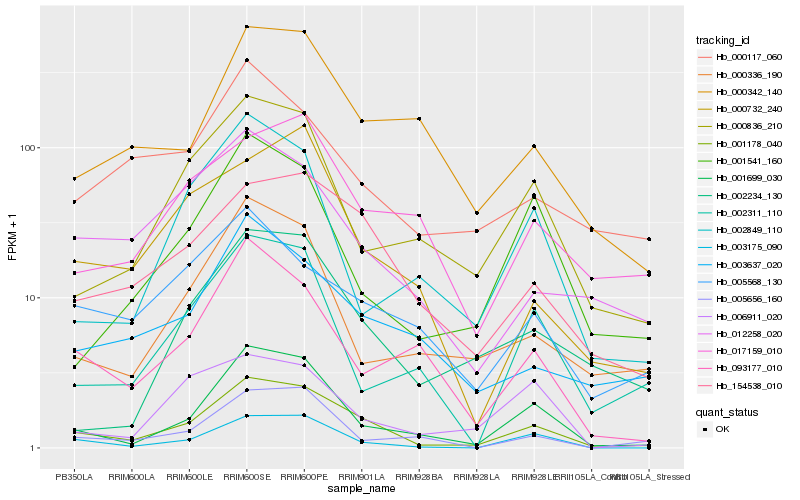
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001178_040 |
0.0 |
desease resistance |
Gene Name: ABC_trans_N |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 2 |
Hb_001699_030 |
0.1503011797 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_154538_010 |
0.1569107145 |
- |
- |
- |
| 4 |
Hb_003175_090 |
0.1695518421 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 5 |
Hb_000836_210 |
0.1946340045 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 6 |
Hb_000342_140 |
0.1997059082 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 7 |
Hb_003637_020 |
0.2018196562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002234_130 |
0.2053287769 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 9 |
Hb_000336_190 |
0.2054201426 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 10 |
Hb_093177_010 |
0.2074620514 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 11 |
Hb_012258_020 |
0.2085204325 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_002311_110 |
0.2108453257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001541_160 |
0.2112466616 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 14 |
Hb_005656_160 |
0.2114468542 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 15 |
Hb_000732_240 |
0.2120107974 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 16 |
Hb_017159_010 |
0.2158060868 |
- |
- |
- |
| 17 |
Hb_005568_130 |
0.2166278563 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_002849_110 |
0.2204667608 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 19 |
Hb_000117_060 |
0.2224410957 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 20 |
Hb_006911_020 |
0.222823783 |
- |
- |
CC-NBS-LRR protein isoform 1 [Theobroma cacao] |