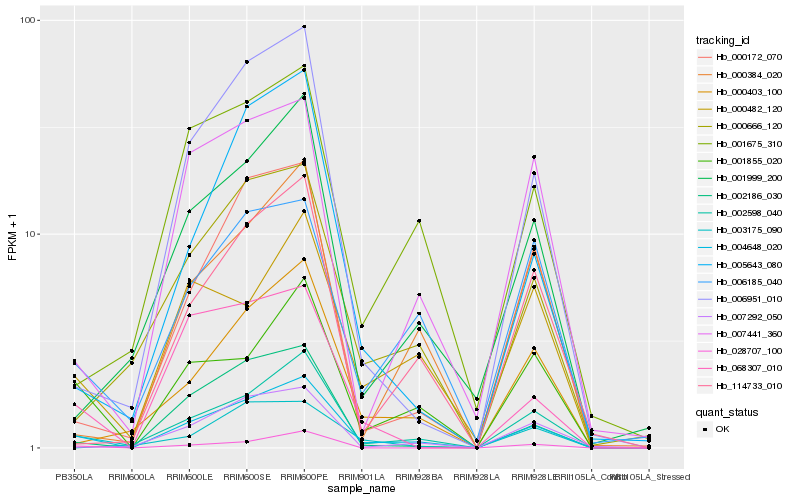
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004648_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 2 |
Hb_001855_020 |
0.1499099526 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_002186_030 |
0.1670908902 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 4 |
Hb_003175_090 |
0.1695919714 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 5 |
Hb_006951_010 |
0.1784969519 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 6 |
Hb_000403_100 |
0.1796042091 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 7 |
Hb_000172_070 |
0.182464262 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_028707_100 |
0.1851577507 |
- |
- |
hypothetical protein RCOM_1687010 [Ricinus communis] |
| 9 |
Hb_000666_120 |
0.1861174134 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_068307_010 |
0.1871757688 |
- |
- |
PREDICTED: phosphomannomutase [Eucalyptus grandis] |
| 11 |
Hb_000482_120 |
0.1873243789 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 12 |
Hb_006185_040 |
0.1873276113 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 13 |
Hb_001999_200 |
0.1880739963 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005643_080 |
0.1886360988 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 15 |
Hb_007292_050 |
0.1893972072 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 16 |
Hb_114733_010 |
0.1898836195 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_002598_040 |
0.1917949855 |
- |
- |
hypothetical protein OsI_22030 [Oryza sativa Indica Group] |
| 18 |
Hb_007441_360 |
0.192272377 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 19 |
Hb_001675_310 |
0.1934904254 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000384_020 |
0.193693802 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |