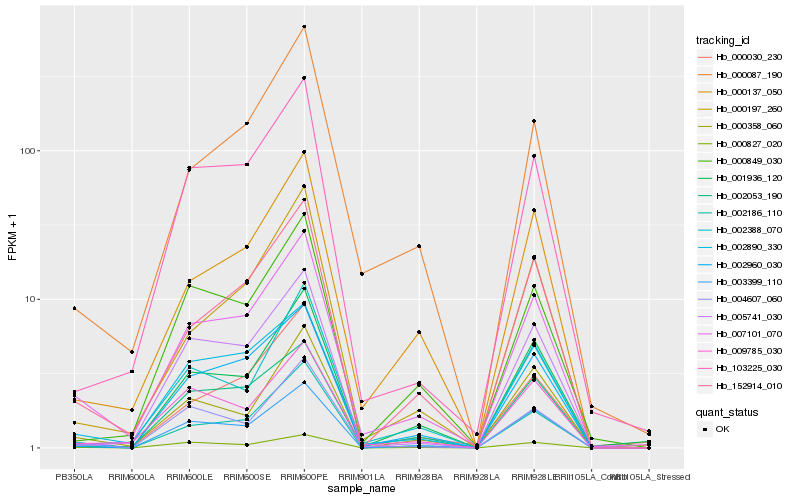
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_070 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 2 |
Hb_003399_110 |
0.0945527802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_152914_010 |
0.1020540547 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 4 |
Hb_002890_330 |
0.102833308 |
- |
- |
caspase, putative [Ricinus communis] |
| 5 |
Hb_103225_030 |
0.1070330277 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_002186_110 |
0.110496535 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 7 |
Hb_000137_050 |
0.111132863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001936_120 |
0.1135250326 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 9 |
Hb_009785_030 |
0.1203162697 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 10 |
Hb_002053_190 |
0.1210107515 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000827_020 |
0.1212223284 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000087_190 |
0.1213592455 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 13 |
Hb_000030_230 |
0.1215604681 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 14 |
Hb_004607_060 |
0.122793474 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 15 |
Hb_000358_060 |
0.1245970858 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 16 |
Hb_002960_030 |
0.1251466237 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 17 |
Hb_000849_030 |
0.1255940462 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 18 |
Hb_005741_030 |
0.1272247334 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 19 |
Hb_000197_260 |
0.1274948715 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 20 |
Hb_002388_070 |
0.1279011404 |
- |
- |
ring finger protein, putative [Ricinus communis] |