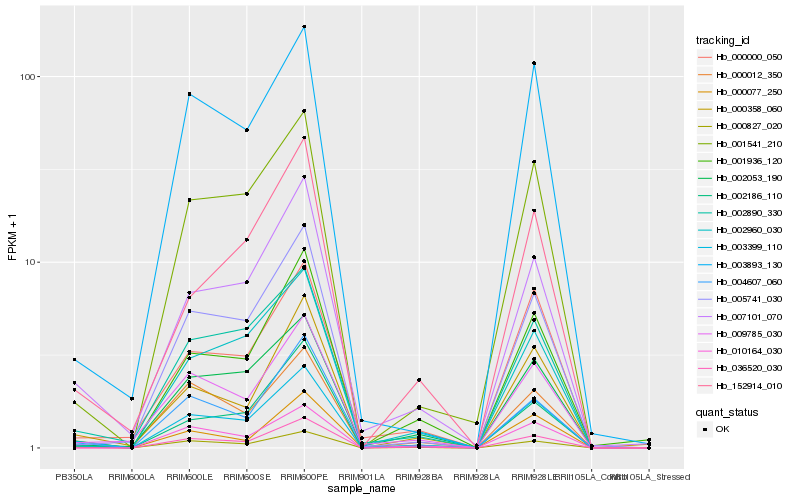
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003399_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000827_020 |
0.0691197852 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_007101_070 |
0.0945527802 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 4 |
Hb_001541_210 |
0.0993457653 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 5 |
Hb_002890_330 |
0.1072096188 |
- |
- |
caspase, putative [Ricinus communis] |
| 6 |
Hb_000358_060 |
0.1095283761 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 7 |
Hb_001936_120 |
0.1140686381 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009785_030 |
0.1144062672 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 9 |
Hb_002053_190 |
0.1172735792 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005741_030 |
0.1195192171 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 11 |
Hb_004607_060 |
0.1200781627 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 12 |
Hb_036520_030 |
0.1212834668 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 13 |
Hb_003893_130 |
0.1243776331 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_152914_010 |
0.1286322675 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 15 |
Hb_002186_110 |
0.1288895584 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 16 |
Hb_000000_050 |
0.12892395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000012_350 |
0.129411879 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 18 |
Hb_000077_250 |
0.1301002391 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 19 |
Hb_002960_030 |
0.1302407636 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 20 |
Hb_010164_030 |
0.1304294764 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |