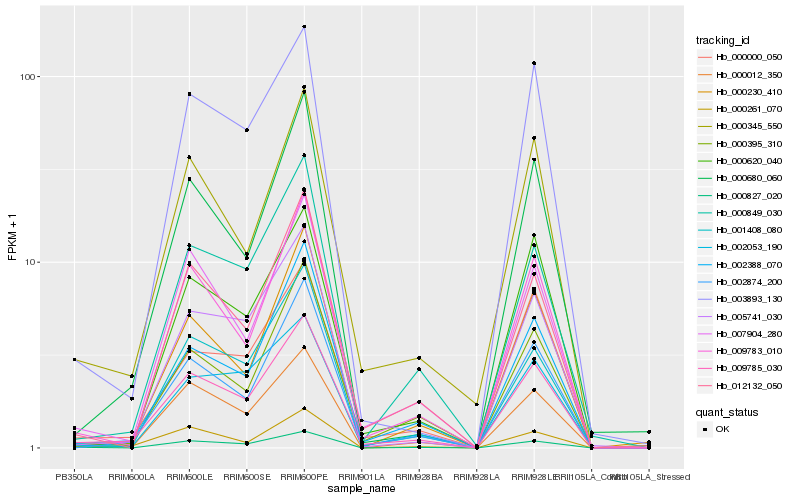
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009785_030 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 2 |
Hb_000345_550 |
0.086661905 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 3 |
Hb_009783_010 |
0.0909341147 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 4 |
Hb_000012_350 |
0.09161877 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 5 |
Hb_002874_200 |
0.0933699457 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 6 |
Hb_007904_280 |
0.0943192179 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 7 |
Hb_012132_050 |
0.0993964164 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 8 |
Hb_000827_020 |
0.1028287966 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_002388_070 |
0.1038528258 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_000395_310 |
0.1040489595 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001408_080 |
0.104971969 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 12 |
Hb_000849_030 |
0.1055833443 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 13 |
Hb_005741_030 |
0.1059566187 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 14 |
Hb_000000_050 |
0.1090364989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000620_040 |
0.1094262414 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000680_060 |
0.110033112 |
- |
- |
hypothetical protein POPTR_0001s20790g [Populus trichocarpa] |
| 17 |
Hb_002053_190 |
0.1127426791 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000261_070 |
0.1132271645 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 19 |
Hb_000230_410 |
0.1135990228 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 20 |
Hb_003893_130 |
0.1141952538 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |