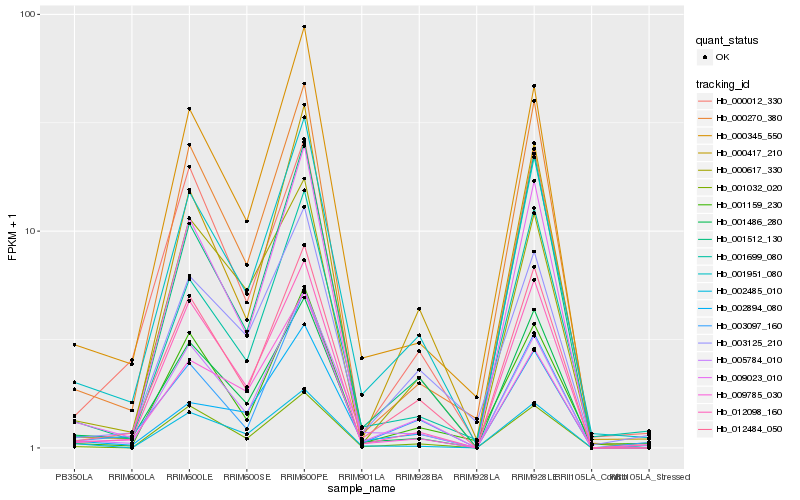
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_080 |
0.0 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 2 |
Hb_012484_050 |
0.0795587476 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 3 |
Hb_005784_010 |
0.080804851 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 4 |
Hb_000345_550 |
0.088279482 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 5 |
Hb_001486_280 |
0.0935818029 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 6 |
Hb_003097_160 |
0.1011682359 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 7 |
Hb_003125_210 |
0.1057904998 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 8 |
Hb_001159_230 |
0.1061896718 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 9 |
Hb_012098_160 |
0.1068660791 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 10 |
Hb_000417_210 |
0.1094031275 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001699_080 |
0.1124197652 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_001512_130 |
0.1127082859 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 13 |
Hb_000270_380 |
0.1127142155 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 14 |
Hb_002894_080 |
0.1143575202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000012_330 |
0.1150014186 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 16 |
Hb_002485_010 |
0.1165461978 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 17 |
Hb_000617_330 |
0.1179271854 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 18 |
Hb_009023_010 |
0.1184549195 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 19 |
Hb_001032_020 |
0.1186391785 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_009785_030 |
0.1190749114 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |