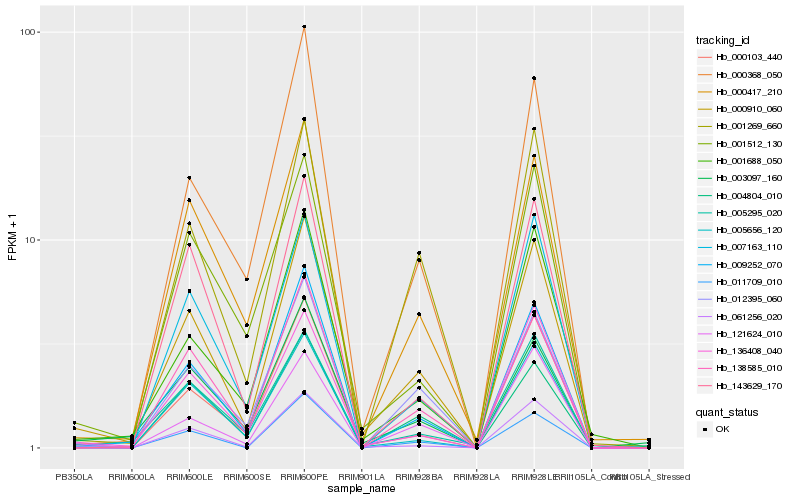
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000910_060 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_009252_070 |
0.1006702262 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 3 |
Hb_001269_660 |
0.1065500637 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 4 |
Hb_012395_060 |
0.1080629249 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 5 |
Hb_000103_440 |
0.1112595119 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 6 |
Hb_136408_040 |
0.1141223953 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 7 |
Hb_005295_020 |
0.1161737819 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 8 |
Hb_001688_050 |
0.1177563478 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 9 |
Hb_138585_010 |
0.1181754248 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 10 |
Hb_121624_010 |
0.1204768854 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 11 |
Hb_007163_110 |
0.1216450556 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 12 |
Hb_003097_160 |
0.1220978136 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 13 |
Hb_000417_210 |
0.1225509735 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_061256_020 |
0.1250509905 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 15 |
Hb_001512_130 |
0.126432517 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 16 |
Hb_011709_010 |
0.1277238175 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 17 |
Hb_000368_050 |
0.1284962274 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 18 |
Hb_005656_120 |
0.1312273433 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 19 |
Hb_004804_010 |
0.1341796645 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 20 |
Hb_143629_170 |
0.1344373107 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |