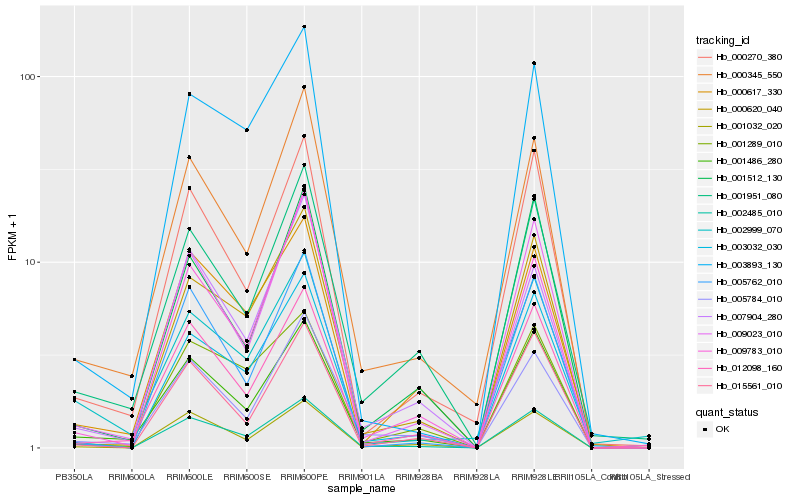
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002485_010 |
0.0 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 2 |
Hb_001486_280 |
0.08920062 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 3 |
Hb_003032_030 |
0.095398337 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001032_020 |
0.0967213791 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 5 |
Hb_009023_010 |
0.105346399 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 6 |
Hb_000345_550 |
0.106615476 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 7 |
Hb_000270_380 |
0.1082291039 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 8 |
Hb_012098_160 |
0.1106308306 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 9 |
Hb_015561_010 |
0.1137505881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000620_040 |
0.1139339135 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_005784_010 |
0.1156510754 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 12 |
Hb_001951_080 |
0.1165461978 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 13 |
Hb_005762_010 |
0.1194934929 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_000617_330 |
0.1199077804 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 15 |
Hb_002999_070 |
0.1220636489 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 16 |
Hb_001289_010 |
0.1229197151 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 17 |
Hb_001512_130 |
0.1255523027 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 18 |
Hb_009783_010 |
0.1260068061 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 19 |
Hb_003893_130 |
0.1280829179 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_007904_280 |
0.1290679449 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |