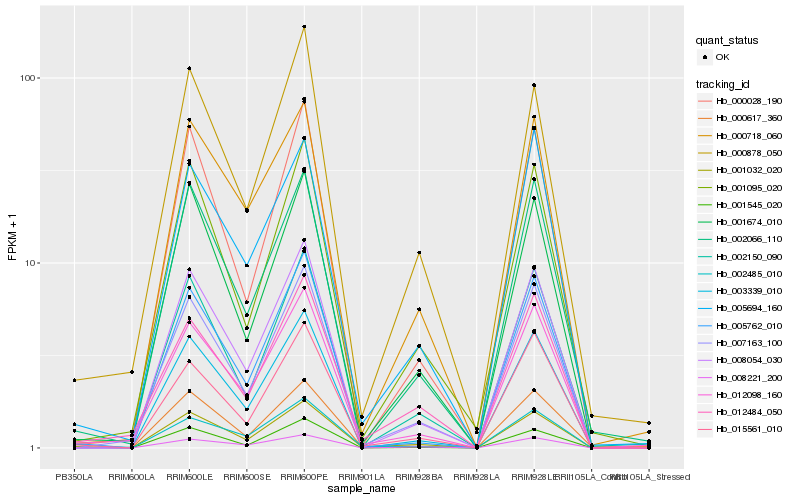
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001032_020 |
0.0 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 2 |
Hb_001674_010 |
0.0796590171 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 3 |
Hb_008054_030 |
0.0873044794 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 4 |
Hb_002066_110 |
0.0874556214 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_012098_160 |
0.0887856289 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 6 |
Hb_007163_100 |
0.0895236693 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 7 |
Hb_015561_010 |
0.0900392549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001095_020 |
0.0941089035 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 9 |
Hb_003339_010 |
0.0945358021 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 10 |
Hb_001545_020 |
0.0946096736 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 11 |
Hb_002150_090 |
0.0966565815 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 12 |
Hb_002485_010 |
0.0967213791 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 13 |
Hb_000617_360 |
0.0971047226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012484_050 |
0.097282591 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 15 |
Hb_000718_060 |
0.097283735 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000878_050 |
0.0974932414 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 17 |
Hb_008221_200 |
0.0991402174 |
- |
- |
PREDICTED: protein trichome birefringence-like 41 [Jatropha curcas] |
| 18 |
Hb_005762_010 |
0.0992803621 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_000028_190 |
0.0998813338 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 20 |
Hb_005694_160 |
0.1018584623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |