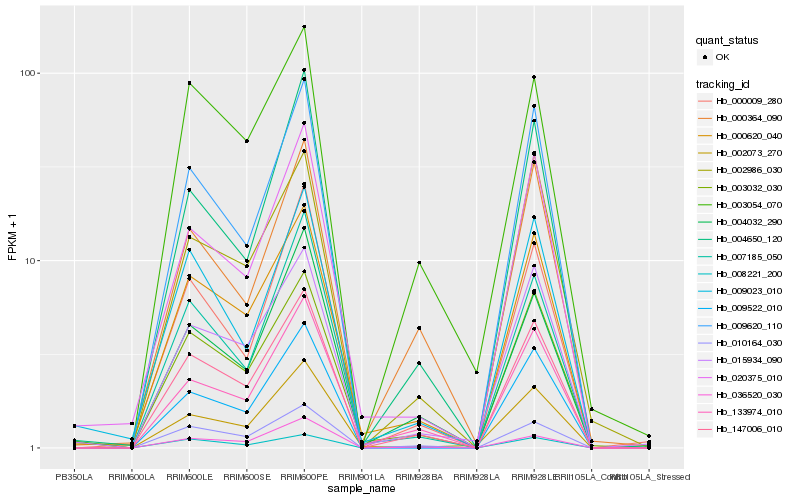
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147006_010 |
0.0 |
- |
- |
receptor like protein 35 [Arabidopsis thaliana] |
| 2 |
Hb_010164_030 |
0.0506433139 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000620_040 |
0.0657379917 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_133974_010 |
0.0788253557 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 5 |
Hb_003032_030 |
0.0799134217 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 6 |
Hb_015934_090 |
0.0891175404 |
- |
- |
PREDICTED: uncharacterized protein LOC105640158 [Jatropha curcas] |
| 7 |
Hb_004032_290 |
0.0914846114 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_003054_070 |
0.0949849817 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_009522_010 |
0.0955766551 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 10 |
Hb_036520_030 |
0.0958048465 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 11 |
Hb_004650_120 |
0.0983991829 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 12 |
Hb_008221_200 |
0.1003584957 |
- |
- |
PREDICTED: protein trichome birefringence-like 41 [Jatropha curcas] |
| 13 |
Hb_002986_030 |
0.1008554344 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 14 |
Hb_009620_110 |
0.1015516939 |
- |
- |
- |
| 15 |
Hb_000009_280 |
0.102081552 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 16 |
Hb_007185_050 |
0.1026943116 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 17 |
Hb_000364_090 |
0.1028068957 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |
| 18 |
Hb_020375_010 |
0.1047117366 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 19 |
Hb_002073_270 |
0.1066761215 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 20 |
Hb_009023_010 |
0.1075954682 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |