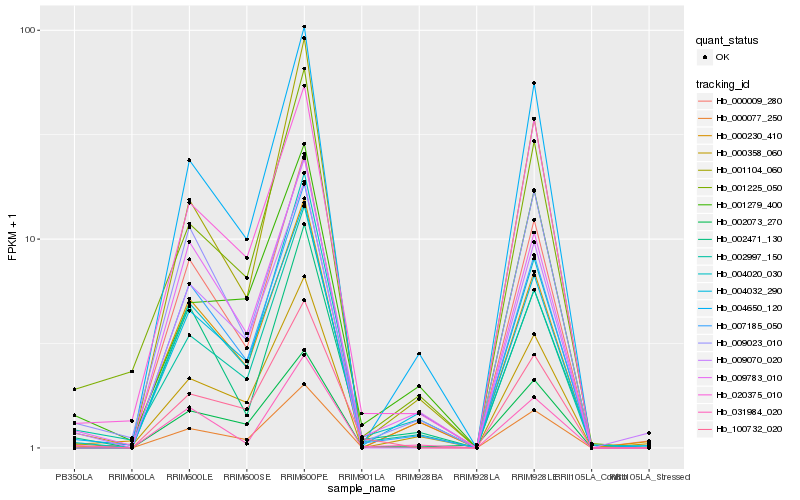
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_250 |
0.0 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 2 |
Hb_002997_150 |
0.0827330721 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 3 |
Hb_020375_010 |
0.0830787855 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 4 |
Hb_004650_120 |
0.0845284651 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 5 |
Hb_007185_050 |
0.0915935797 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 6 |
Hb_004020_030 |
0.0923509956 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 7 |
Hb_009070_020 |
0.0956558646 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 8 |
Hb_001225_050 |
0.0971797861 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 9 |
Hb_000358_060 |
0.0996195376 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 10 |
Hb_009783_010 |
0.1012528947 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 11 |
Hb_000009_280 |
0.1035639772 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 12 |
Hb_001104_060 |
0.1064881156 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 13 |
Hb_100732_020 |
0.1074426883 |
- |
- |
hypothetical protein POPTR_0001s15930g [Populus trichocarpa] |
| 14 |
Hb_009023_010 |
0.1087174421 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 15 |
Hb_002471_130 |
0.1092151263 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 16 |
Hb_002073_270 |
0.1094472643 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 17 |
Hb_004032_290 |
0.11130822 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000230_410 |
0.1116454009 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 19 |
Hb_031984_020 |
0.1116778613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001279_400 |
0.1121440319 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |