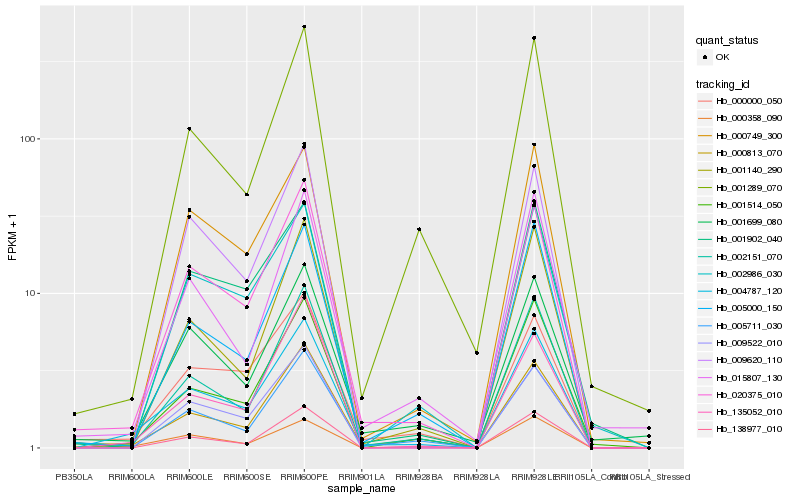
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004787_120 |
0.0 |
- |
- |
PREDICTED: transcription factor TCP14 [Jatropha curcas] |
| 2 |
Hb_020375_010 |
0.0858283805 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 3 |
Hb_000813_070 |
0.098188582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000358_090 |
0.1013993073 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_001514_050 |
0.1050267051 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 6 |
Hb_002151_070 |
0.1083791109 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 2-like [Jatropha curcas] |
| 7 |
Hb_000000_050 |
0.1159671182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005711_030 |
0.1176833131 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 9 |
Hb_005000_150 |
0.119111828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000749_300 |
0.1197515755 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 11 |
Hb_135052_010 |
0.1248619861 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_001140_290 |
0.1254063166 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 13 |
Hb_009522_010 |
0.1255694908 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 14 |
Hb_138977_010 |
0.1265027494 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_009620_110 |
0.1278567306 |
- |
- |
- |
| 16 |
Hb_001289_070 |
0.1303419303 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 17 |
Hb_002986_030 |
0.131441666 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 18 |
Hb_001699_080 |
0.1323676547 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_001902_040 |
0.1342657727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_015807_130 |
0.1347036733 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |