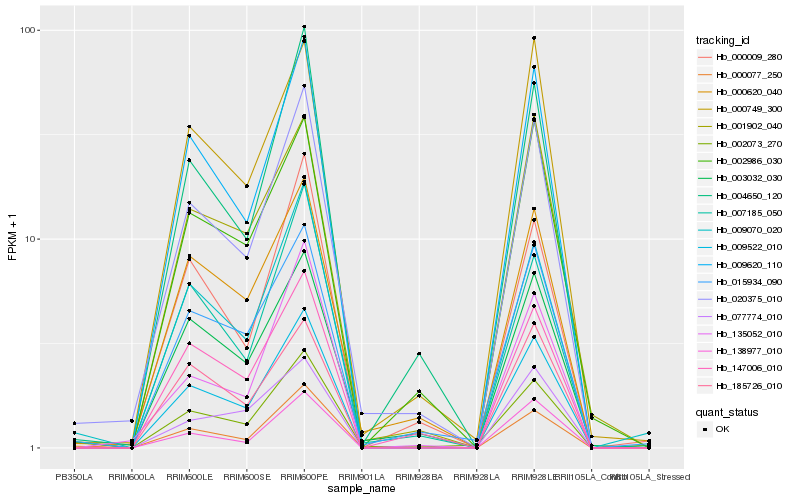
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009522_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 2 |
Hb_009620_110 |
0.0404801028 |
- |
- |
- |
| 3 |
Hb_020375_010 |
0.0857390422 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 4 |
Hb_002073_270 |
0.0911638549 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 5 |
Hb_185726_010 |
0.0915931735 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 6 |
Hb_009070_020 |
0.0952741063 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 7 |
Hb_147006_010 |
0.0955766551 |
- |
- |
receptor like protein 35 [Arabidopsis thaliana] |
| 8 |
Hb_004650_120 |
0.0960357597 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 9 |
Hb_138977_010 |
0.0989686967 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000749_300 |
0.1021767321 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 11 |
Hb_077774_010 |
0.1043600359 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 12 |
Hb_015934_090 |
0.1065730546 |
- |
- |
PREDICTED: uncharacterized protein LOC105640158 [Jatropha curcas] |
| 13 |
Hb_000620_040 |
0.1084534879 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001902_040 |
0.1086584739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007185_050 |
0.1095354066 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 16 |
Hb_002986_030 |
0.1105017172 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 17 |
Hb_000009_280 |
0.111829949 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 18 |
Hb_003032_030 |
0.1145383415 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000077_250 |
0.1145534146 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 20 |
Hb_135052_010 |
0.1152430289 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |