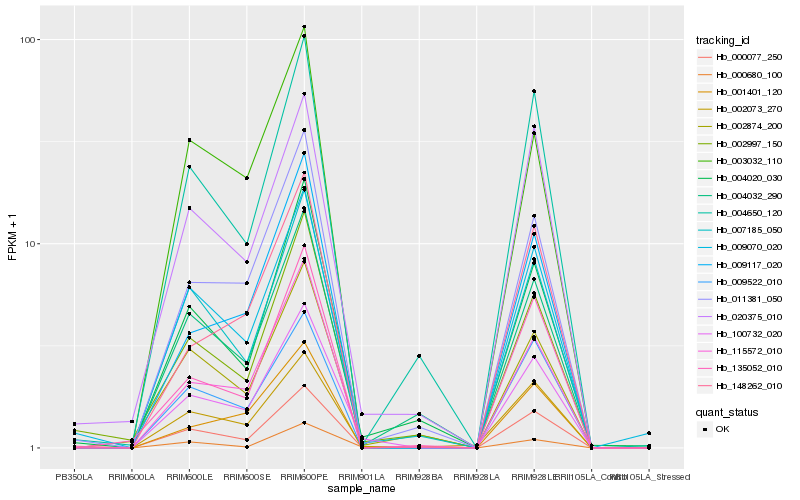
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100732_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s15930g [Populus trichocarpa] |
| 2 |
Hb_002073_270 |
0.1030593257 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 3 |
Hb_011381_050 |
0.105952943 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 4 |
Hb_007185_050 |
0.1071957323 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 5 |
Hb_000077_250 |
0.1074426883 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 6 |
Hb_115572_010 |
0.1080240222 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 7 |
Hb_009522_010 |
0.1163908679 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 8 |
Hb_004020_030 |
0.1178502514 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 9 |
Hb_009117_020 |
0.1196852723 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 10 |
Hb_020375_010 |
0.1197451681 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 11 |
Hb_009070_020 |
0.1238637778 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 12 |
Hb_004650_120 |
0.1247007851 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 13 |
Hb_000680_100 |
0.1263859319 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_001401_120 |
0.1273675243 |
- |
- |
hypothetical protein JCGZ_04273 [Jatropha curcas] |
| 15 |
Hb_135052_010 |
0.1276946806 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_002997_150 |
0.127829194 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 17 |
Hb_004032_290 |
0.127925527 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_003032_110 |
0.1303227582 |
- |
- |
- |
| 19 |
Hb_148262_010 |
0.1303595422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002874_200 |
0.1308203937 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |