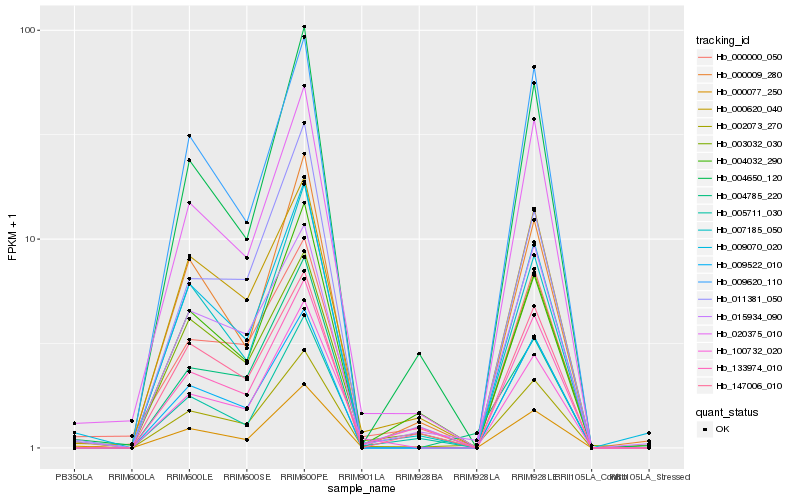
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_270 |
0.0 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 2 |
Hb_009522_010 |
0.0911638549 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 3 |
Hb_020375_010 |
0.0932603237 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 4 |
Hb_015934_090 |
0.095494927 |
- |
- |
PREDICTED: uncharacterized protein LOC105640158 [Jatropha curcas] |
| 5 |
Hb_100732_020 |
0.1030593257 |
- |
- |
hypothetical protein POPTR_0001s15930g [Populus trichocarpa] |
| 6 |
Hb_007185_050 |
0.1037497286 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 7 |
Hb_000620_040 |
0.1046408969 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_009620_110 |
0.1050177542 |
- |
- |
- |
| 9 |
Hb_004650_120 |
0.1058455557 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 10 |
Hb_133974_010 |
0.1061768415 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 11 |
Hb_147006_010 |
0.1066761215 |
- |
- |
receptor like protein 35 [Arabidopsis thaliana] |
| 12 |
Hb_000077_250 |
0.1094472643 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 13 |
Hb_003032_030 |
0.1153650527 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000009_280 |
0.1185852182 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 15 |
Hb_004032_290 |
0.1189251698 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_009070_020 |
0.120814285 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 17 |
Hb_004785_220 |
0.1217282076 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.2 [Populus euphratica] |
| 18 |
Hb_005711_030 |
0.1220709654 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 19 |
Hb_011381_050 |
0.1252995308 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 20 |
Hb_000000_050 |
0.1276466232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |