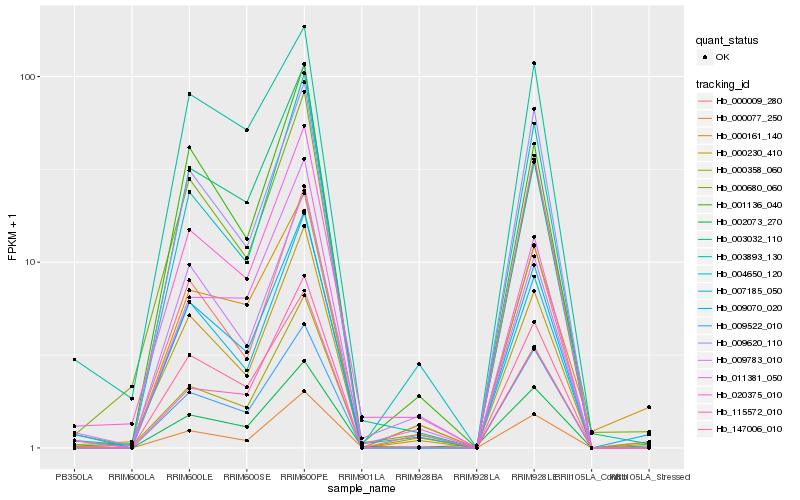
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009070_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 2 |
Hb_000680_060 |
0.0923146929 |
- |
- |
hypothetical protein POPTR_0001s20790g [Populus trichocarpa] |
| 3 |
Hb_009522_010 |
0.0952741063 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 4 |
Hb_000077_250 |
0.0956558646 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 5 |
Hb_000009_280 |
0.0963957065 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 6 |
Hb_000161_140 |
0.0989519578 |
- |
- |
PREDICTED: WEB family protein At3g51220 [Jatropha curcas] |
| 7 |
Hb_007185_050 |
0.0995355625 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 8 |
Hb_000230_410 |
0.1006959934 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 9 |
Hb_009620_110 |
0.1037604581 |
- |
- |
- |
| 10 |
Hb_001136_040 |
0.10885366 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_004650_120 |
0.1101346395 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 12 |
Hb_000358_060 |
0.1114630522 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 13 |
Hb_009783_010 |
0.1146028717 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 14 |
Hb_020375_010 |
0.1162713989 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 15 |
Hb_003893_130 |
0.118781523 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_115572_010 |
0.1195502068 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 17 |
Hb_002073_270 |
0.120814285 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 18 |
Hb_003032_110 |
0.1215825965 |
- |
- |
- |
| 19 |
Hb_011381_050 |
0.1225275426 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 20 |
Hb_147006_010 |
0.1235922327 |
- |
- |
receptor like protein 35 [Arabidopsis thaliana] |