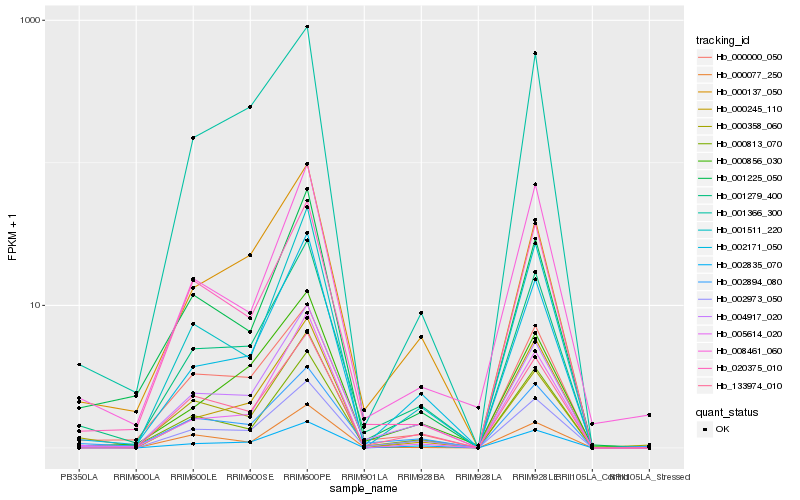
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_400 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |
| 2 |
Hb_004917_020 |
0.0634861898 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 3 |
Hb_002973_050 |
0.0817295034 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 4 |
Hb_000813_070 |
0.0855043263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002171_050 |
0.0861719317 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 6 |
Hb_002894_080 |
0.0891583975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000000_050 |
0.0895683036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_133974_010 |
0.0963005847 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 9 |
Hb_002835_070 |
0.0976900726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000245_110 |
0.10046301 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 11 |
Hb_000137_050 |
0.1005337122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_020375_010 |
0.1044319524 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 13 |
Hb_000358_060 |
0.1105457802 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 14 |
Hb_000077_250 |
0.1121440319 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 15 |
Hb_001511_220 |
0.1122800027 |
- |
- |
hypothetical protein POPTR_0008s06990g [Populus trichocarpa] |
| 16 |
Hb_001225_050 |
0.1124481315 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 17 |
Hb_001366_300 |
0.11315593 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 18 |
Hb_008461_060 |
0.1132197993 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 19 |
Hb_005614_020 |
0.113729365 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000856_030 |
0.1162795873 |
- |
- |
ATP binding protein, putative [Ricinus communis] |